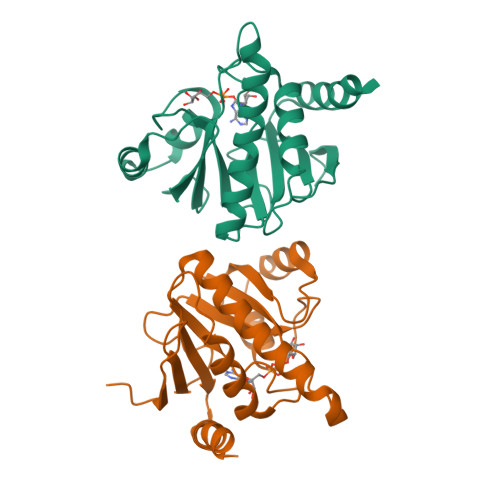
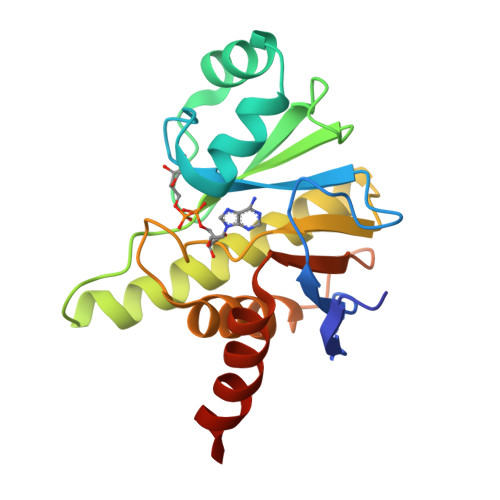
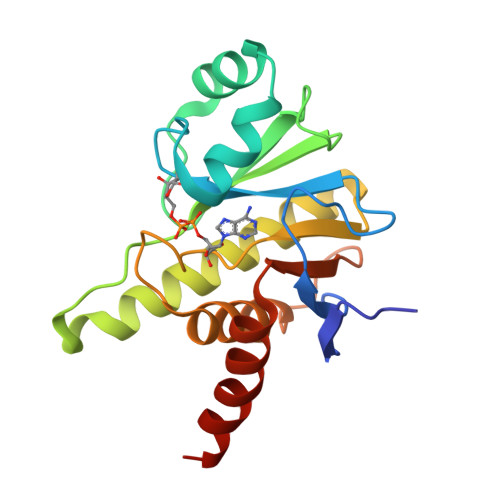
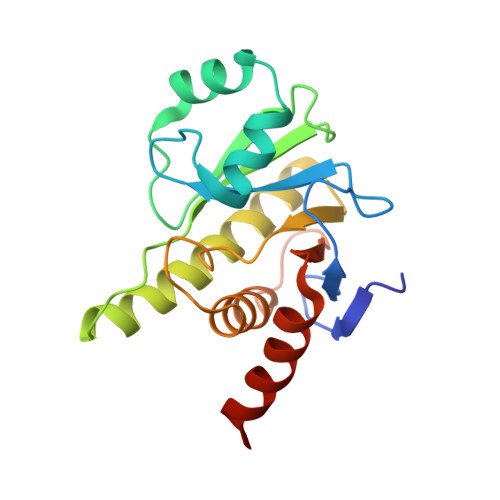
Crystal structure and biochemical activity of the macrodomain from rubella virus p150.
Stoll, G.A., Nikolopoulos, N., Zhai, H., Zhang, L., Douse, C.H., Modis, Y.(2024) J Virol 98: e0177723-e0177723
- PubMed: 38289106
- DOI: https://doi.org/10.1128/jvi.01777-23
- Primary Citation of Related Structures:
8P0C, 8P0E - PubMed Abstract:
Rubella virus encodes a nonstructural polyprotein with RNA polymerase, methyltransferase, and papain-like cysteine protease activities, along with a putative macrodomain of unknown function. Macrodomains bind ADP-ribose adducts, a post-translational modification that plays a key role in host-virus conflicts. Some macrodomains can also remove the mono-ADP-ribose adduct or degrade poly-ADP-ribose chains. Here, we report high-resolution crystal structures of the macrodomain from rubella virus nonstructural protein p150, with and without ADP-ribose binding. The overall fold is most similar to macroD-type macrodomains from various nonviral species. The specific composition and structure of the residues that coordinate ADP-ribose in the rubella virus macrodomain are most similar to those of macrodomains from alphaviruses. Isothermal calorimetry shows that the rubella virus macrodomain binds ADP-ribose in solution. Enzyme assays show that the rubella virus macrodomain can hydrolyze both mono- and poly-ADP-ribose adducts. Site-directed mutagenesis identifies Asn39 and Cys49 required for mono-ADP-ribosylhydrolase (de-MARylation) activity.IMPORTANCERubella virus remains a global health threat. Rubella infections during pregnancy can cause serious congenital pathology, for which no antiviral treatments are available. Our work demonstrates that, like alpha- and coronaviruses, rubiviruses encode a mono-ADP-ribosylhydrolase with a structurally conserved macrodomain fold to counteract MARylation by poly (ADP-ribose) polymerases (PARPs) in the host innate immune response. Our structural data will guide future efforts to develop novel antiviral therapeutics against rubella or infections with related viruses.
Organizational Affiliation:
Molecular Immunity Unit, Department of Medicine, University of Cambridge, MRC Laboratory of Molecular Biology, Cambridge, United Kingdom.