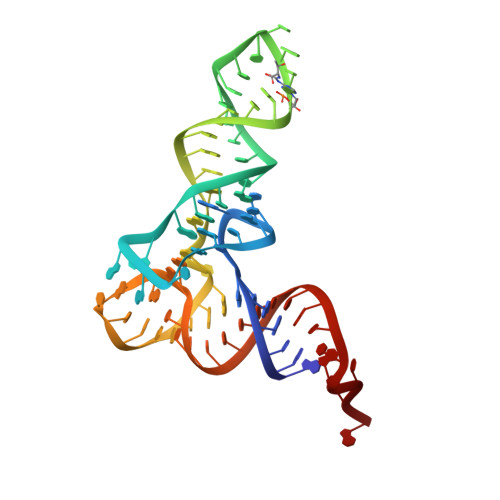
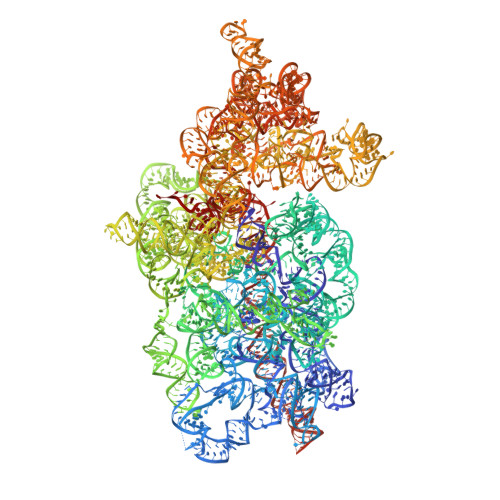
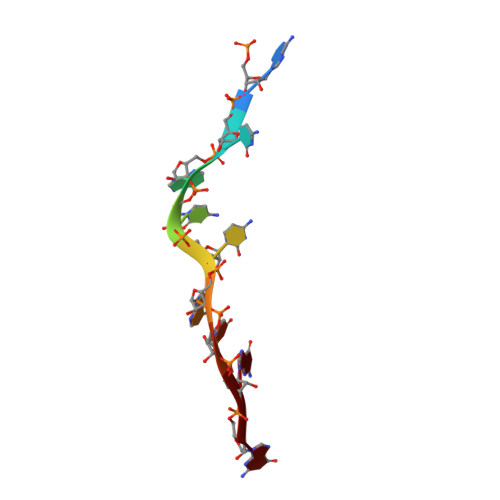
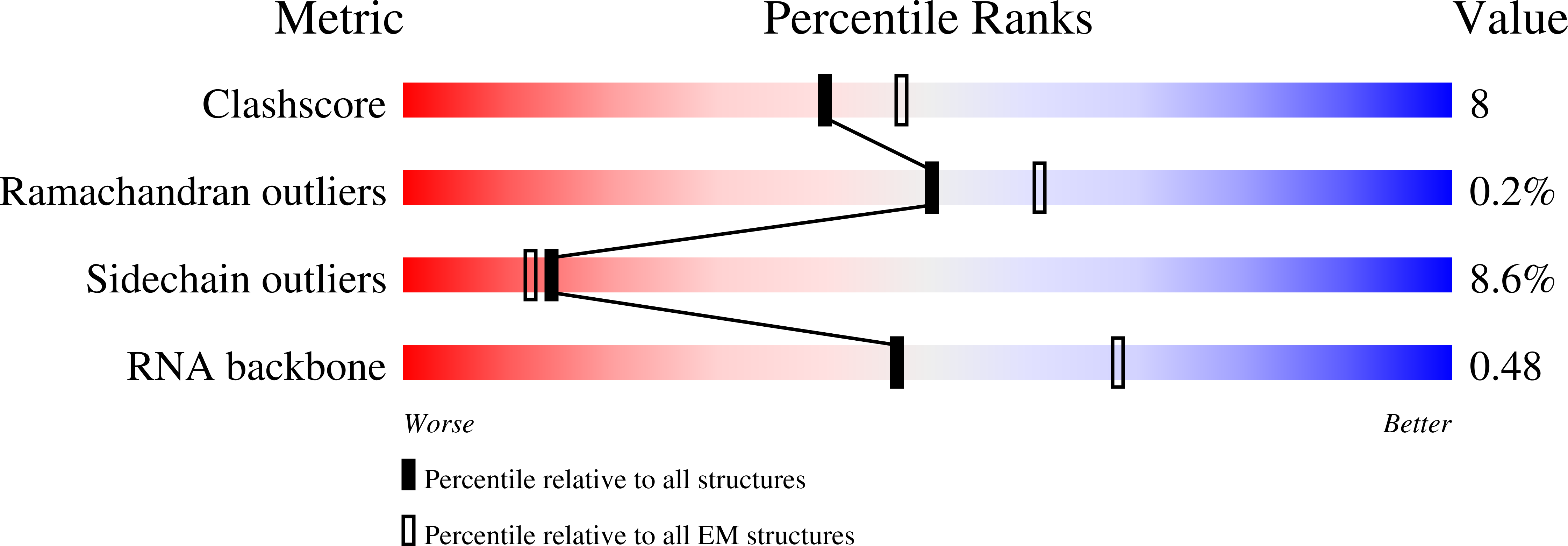N 6 -methyladenosine in 5' UTR does not promote translation initiation.
Guca, E., Alarcon, R., Palo, M.Z., Santos, L., Alonso-Gil, S., Davyt, M., de Lima, L.H.F., Boissier, F., Das, S., Zagrovic, B., Puglisi, J.D., Hashem, Y., Ignatova, Z.(2024) Mol Cell 84: 584-595.e6
- PubMed: 38244546
- DOI: https://doi.org/10.1016/j.molcel.2023.12.028
- Primary Citation of Related Structures:
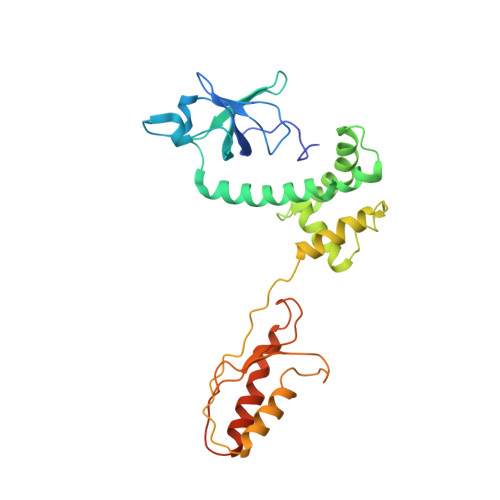
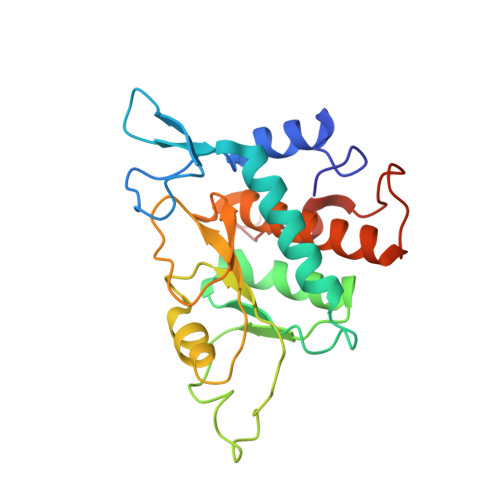
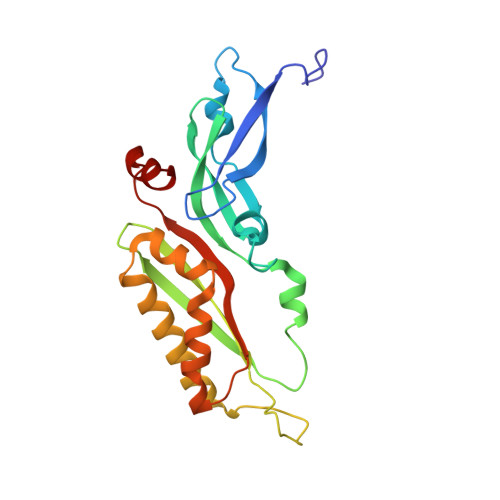
8P03, 8P09 - PubMed Abstract:
The most abundant N 6 -methyladenosine (m 6 A) modification on mRNAs is installed non-stoichiometrically across transcripts, with 5' untranslated regions (5' UTRs) being the least conductive. 5' UTRs are essential for translation initiation, yet the molecular mechanisms orchestrated by m 6 A remain poorly understood. Here, we combined structural, biochemical, and single-molecule approaches and show that at the most common position, a single m 6 A does not affect translation yields, the kinetics of translation initiation complex assembly, or start codon recognition both under permissive growth and following exposure to oxidative stress. Cryoelectron microscopy (cryo-EM) structures of the late preinitiation complex reveal that m 6 A purine ring established stacking interactions with an arginine side chain of the initiation factor eIF2α, although with only a marginal energy contribution, as estimated computationally. These findings provide molecular insights into m 6 A interactions with the initiation complex and suggest that the subtle stabilization is unlikely to affect the translation dynamics under homeostatic conditions or stress.
Organizational Affiliation:
INSERM U1212 Acides nucléiques: Régulations Naturelle et Artificielle (ARNA), Institut Européen de Chimie et Biologie, Université de Bordeaux, Pessac 33607, France.