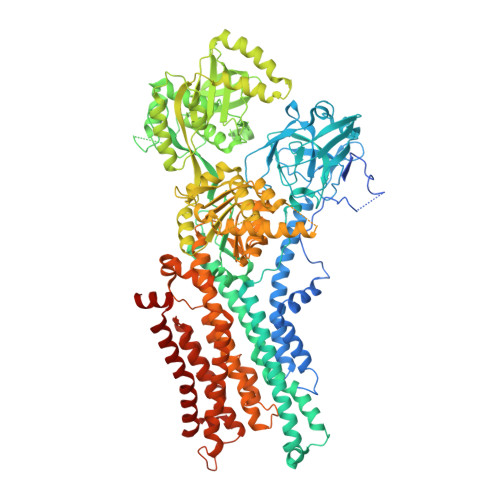
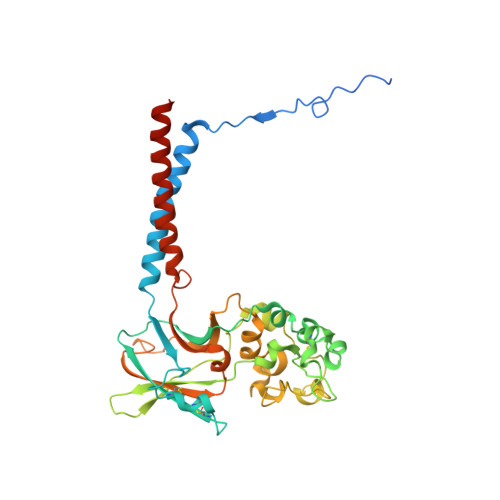
Activation and substrate specificity of the human P4-ATPase ATP8B1.
Dieudonne, T., Kummerer, F., Laursen, M.J., Stock, C., Flygaard, R.K., Khalid, S., Lenoir, G., Lyons, J.A., Lindorff-Larsen, K., Nissen, P.(2023) Nat Commun 14: 7492-7492
- PubMed: 37980352
- DOI: https://doi.org/10.1038/s41467-023-42828-9
- Primary Citation of Related Structures:
8OX4, 8OX5, 8OX6, 8OX7, 8OX8, 8OX9, 8OXA, 8OXB, 8OXC - PubMed Abstract:
Asymmetric distribution of phospholipids in eukaryotic membranes is essential for cell integrity, signaling pathways, and vesicular trafficking. P4-ATPases, also known as flippases, participate in creating and maintaining this asymmetry through active transport of phospholipids from the exoplasmic to the cytosolic leaflet. Here, we present a total of nine cryo-electron microscopy structures of the human flippase ATP8B1-CDC50A complex at 2.4 to 3.1 Å overall resolution, along with functional and computational studies, addressing the autophosphorylation steps from ATP, substrate recognition and occlusion, as well as a phosphoinositide binding site. We find that the P4-ATPase transport site is occupied by water upon phosphorylation from ATP. Additionally, we identify two different autoinhibited states, a closed and an outward-open conformation. Furthermore, we identify and characterize the PI(3,4,5)P 3 binding site of ATP8B1 in an electropositive pocket between transmembrane segments 5, 7, 8, and 10. Our study also highlights the structural basis of a broad lipid specificity of ATP8B1 and adds phosphatidylinositol as a transport substrate for ATP8B1. We report a critical role of the sn-2 ester bond of glycerophospholipids in substrate recognition by ATP8B1 through conserved S403. These findings provide fundamental insights into ATP8B1 catalytic cycle and regulation, and substrate recognition in P4-ATPases.
- DANDRITE, Nordic EMBL Partnership for Molecular Medicine, Department of Molecular Biology and Genetics, Aarhus University, Aarhus, Denmark. thibaud.dieudonne@i2bc.paris-saclay.fr.
Organizational Affiliation: