Structural and functional studies of geldanamycin amide synthase ShGdmF
Ewert, W., Bartens, C., Heutling, A., Ongouta, J., Vogt, M., Kishore, A., Zeilinger, C., Preller, M.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| GdmF | 263 | Streptomyces hygroscopicus | Mutation(s): 0 Gene Names: gdmF | 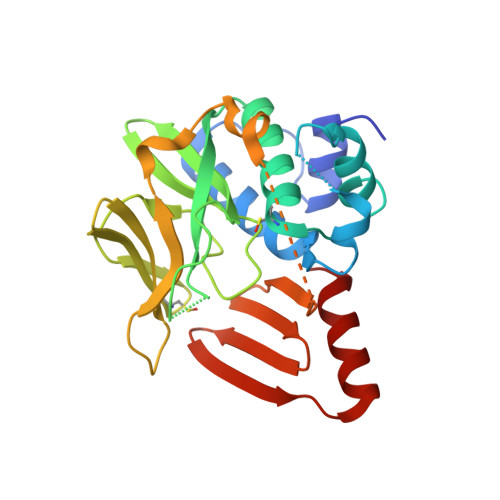 | |
UniProt | |||||
Find proteins for Q84G21 (Streptomyces hygroscopicus) Explore Q84G21 Go to UniProtKB: Q84G21 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q84G21 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| W0E (Subject of Investigation/LOI) Query on W0E | F [auth A] | 3-azanyl-5-methyl-phenol C7 H9 N O NCUABBHFJSFKOJ-UHFFFAOYSA-N |  | ||
| PEG Query on PEG | Q [auth A] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| SO4 Query on SO4 | K [auth A], S [auth A] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| EDO Query on EDO | B [auth A] G [auth A] H [auth A] I [auth A] M [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| ACT Query on ACT | C [auth A] D [auth A] E [auth A] J [auth A] L [auth A] | ACETATE ION C2 H3 O2 QTBSBXVTEAMEQO-UHFFFAOYSA-M |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| CSO Query on CSO | A | L-PEPTIDE LINKING | C3 H7 N O3 S |  | CYS |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 72.544 | α = 90 |
| b = 96.39 | β = 90 |
| c = 86.631 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| Coot | model building |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |