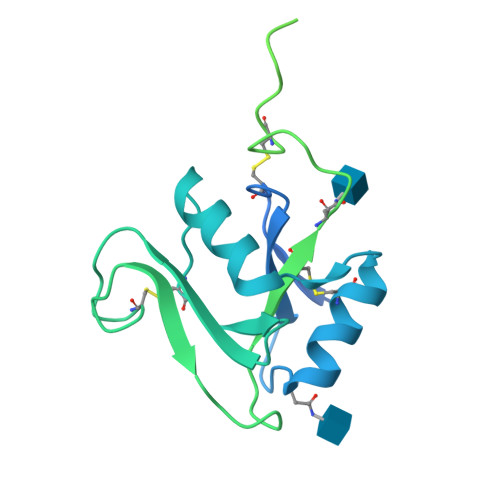
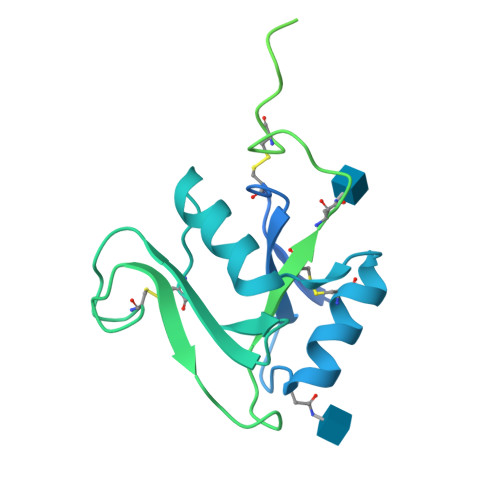
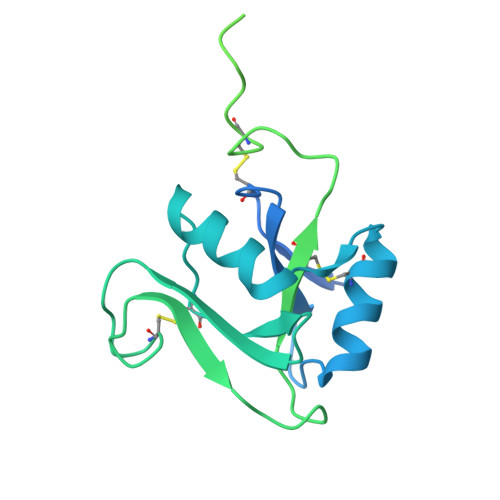
Structure of the murine LYVE-1 (lymphatic vessel endothelial receptor-1) hyaluronan binding domain in an unliganded state
Bano, F., Banerji, S., Ni, T., Green, D.E., DeAngelis, P.L., Paci, E., Lepsik, M., Jackson, D.J., Gilbert, R.J.C.(2025) Nature Communications