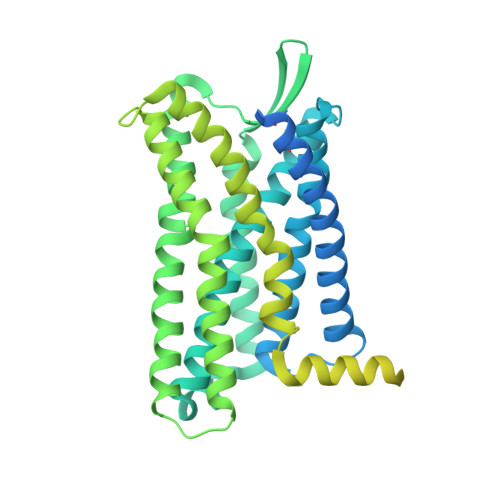
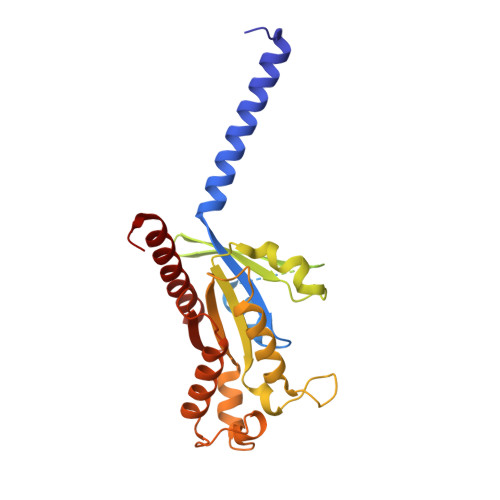
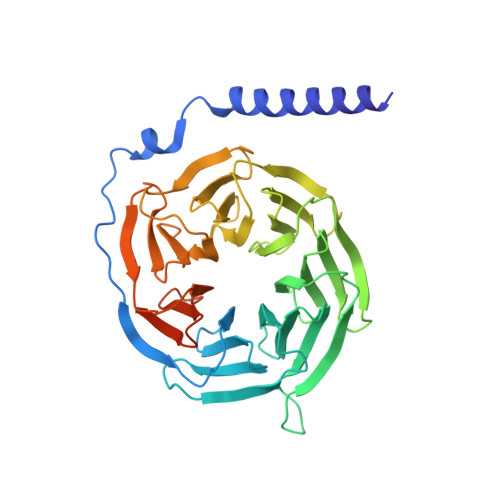
Unveiling the structural mechanisms of nonpeptide ligand recognition and activation in human chemokine receptor CCR8.
Jiang, S., Lin, X., Wu, L., Wang, L., Wu, Y., Xu, Z., Xu, F.(2024) Sci Adv 10: eadj7500-eadj7500
- PubMed: 38306437
- DOI: https://doi.org/10.1126/sciadv.adj7500
- Primary Citation of Related Structures:
8KFX, 8KFY, 8KFZ - PubMed Abstract:
The human CC chemokine receptor 8 (CCR8) is an emerging therapeutic target for cancer immunotherapy and autoimmune diseases. Understanding the molecular recognition of CCR8, particularly with nonpeptide ligands, is valuable for drug development. Here, we report three cryo-electron microscopy structures of human CCR8 complexed with G i trimers in the ligand-free state or activated by nonpeptide agonists LMD-009 and ZK 756326. A conserved Y 1.39 Y 3.32 E 7.39 motif in the orthosteric binding pocket is shown to play a crucial role in the chemokine and nonpeptide ligand recognition. Structural and functional analyses indicate that the lack of conservation in Y114 3.33 and Y172 4.64 among the CC chemokine receptors could potentially contribute to the selectivity of the nonpeptide ligand binding to CCR8. These findings present the characterization of the molecular interaction between a nonpeptide agonist and a chemokine receptor, aiding the development of therapeutics targeting related diseases through a structure-based approach.
Organizational Affiliation:
iHuman Institute, ShanghaiTech University, Pudong, Shanghai 201210, China.