Structural basis for EROS binding to human phagocyte NADPH oxidase NOX2.
Liang, S., Liu, A., Liu, Y., Wang, F., Zhou, Y., Long, Y., Wang, T., Liu, Z., Ren, R., Ye, R.D.(2024) Proc Natl Acad Sci U S A 121: e2320388121-e2320388121
- PubMed: 38805284
- DOI: https://doi.org/10.1073/pnas.2320388121
- Primary Citation of Related Structures:
8KEI - PubMed Abstract:
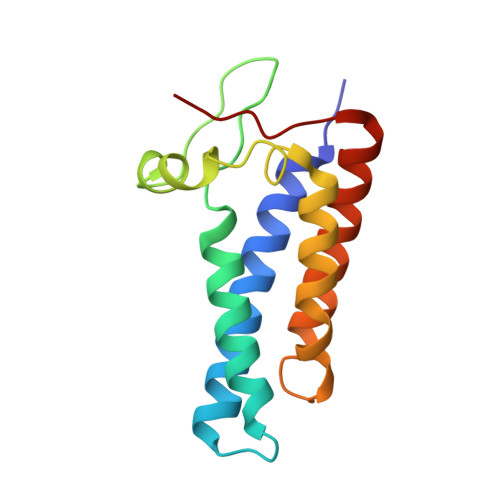
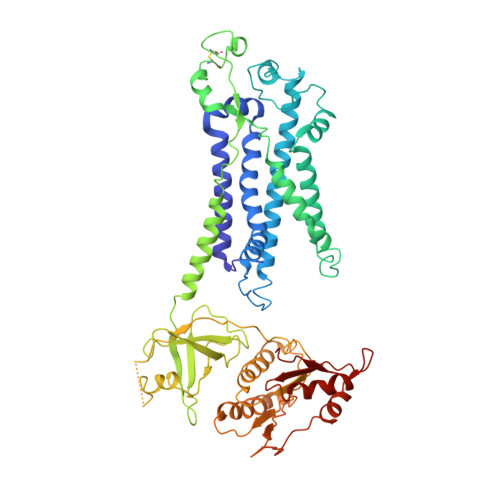
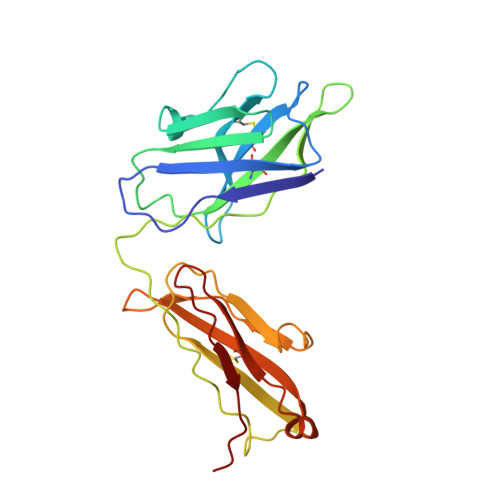
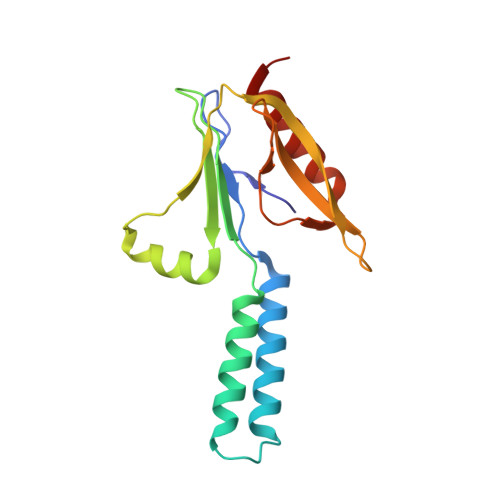
Essential for reactive oxygen species (EROS) protein is a recently identified molecular chaperone of NOX2 (gp91 phox ), the catalytic subunit of phagocyte NADPH oxidase. Deficiency in EROS is a recently identified cause for chronic granulomatous disease, a genetic disorder with recurrent bacterial and fungal infections. Here, we report a cryo-EM structure of the EROS-NOX2-p22 phox heterotrimeric complex at an overall resolution of 3.56Å. EROS and p22 phox are situated on the opposite sides of NOX2, and there is no direct contact between them. EROS associates with NOX2 through two antiparallel transmembrane (TM) α-helices and multiple β-strands that form hydrogen bonds with the cytoplasmic domain of NOX2. EROS binding induces a 79° upward bend of TM2 and a 48° backward rotation of the lower part of TM6 in NOX2, resulting in an increase in the distance between the two hemes and a shift of the binding site for flavin adenine dinucleotide (FAD). These conformational changes are expected to compromise superoxide production by NOX2, suggesting that the EROS-bound NOX2 is in a protected state against activation. Phorbol myristate acetate, an activator of NOX2 in vitro, is able to induce dissociation of NOX2 from EROS with concurrent increase in FAD binding and superoxide production in a transfected COS-7 model. In differentiated neutrophil-like HL-60, the majority of NOX2 on the cell surface is dissociated with EROS. Further studies are required to delineate how EROS dissociates from NOX2 during its transport to cell surface, which may be a potential mechanism for regulation of NOX2 activation.
Organizational Affiliation:
Kobilka Institute of Innovative Drug Discovery, School of Medicine, The Chinese University of Hong Kong, Shenzhen, Guangdong 518172, China.