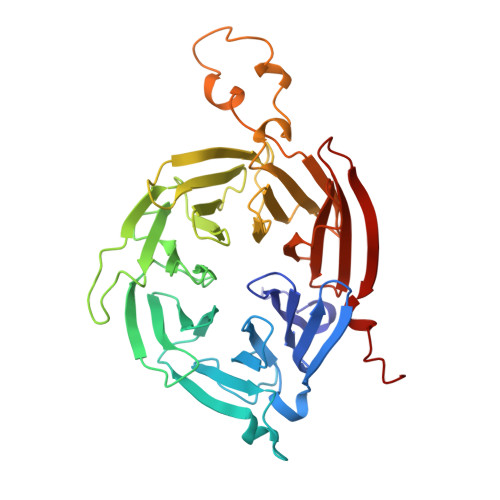
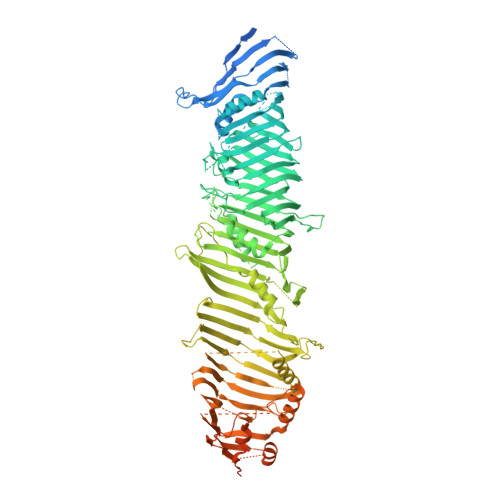
Structural basis for lipid transfer by the ATG2A-ATG9A complex.
Wang, Y., Dahmane, S., Ti, R., Mai, X., Zhu, L., Carlson, L.A., Stjepanovic, G.(2024) Nat Struct Mol Biol
- PubMed: 39174844
- DOI: https://doi.org/10.1038/s41594-024-01376-6
- Primary Citation of Related Structures:
8KBX, 8KBY, 8KBZ, 8KC3, 8Y1L - PubMed Abstract:
Autophagy is characterized by the formation of double-membrane vesicles called autophagosomes. Autophagy-related proteins (ATGs) 2A and 9A have an essential role in autophagy by mediating lipid transfer and re-equilibration between membranes for autophagosome formation. Here we report the cryo-electron microscopy structures of human ATG2A in complex with WD-repeat protein interacting with phosphoinositides 4 (WIPI4) at 3.2 Å and the ATG2A-WIPI4-ATG9A complex at 7 Å global resolution. On the basis of molecular dynamics simulations, we propose a mechanism of lipid extraction from the donor membranes. Our analysis revealed 3:1 stoichiometry of the ATG9A-ATG2A complex, directly aligning the ATG9A lateral pore with ATG2A lipid transfer cavity, and an interaction of the ATG9A trimer with both the N-terminal and the C-terminal tip of rod-shaped ATG2A. Cryo-electron tomography of ATG2A liposome-binding states showed that ATG2A tethers lipid vesicles at different orientations. In summary, this study provides a molecular basis for the growth of the phagophore membrane and lends structural insights into spatially coupled lipid transport and re-equilibration during autophagosome formation.
Organizational Affiliation:
Kobilka Institute of Innovative Drug Discovery, School of Medicine, The Chinese University of Hong Kong, Shenzhen, Shenzhen, China.