Structural and dynamic insights into the activation of the mu-opioid receptor by an allosteric modulator.
Kaneko, S., Imai, S., Uchikubo-Kamo, T., Hisano, T., Asao, N., Shirouzu, M., Shimada, I.(2024) Nat Commun 15: 3544-3544
- PubMed: 38740791
- DOI: https://doi.org/10.1038/s41467-024-47792-6
- Primary Citation of Related Structures:
8K9K, 8K9L - PubMed Abstract:
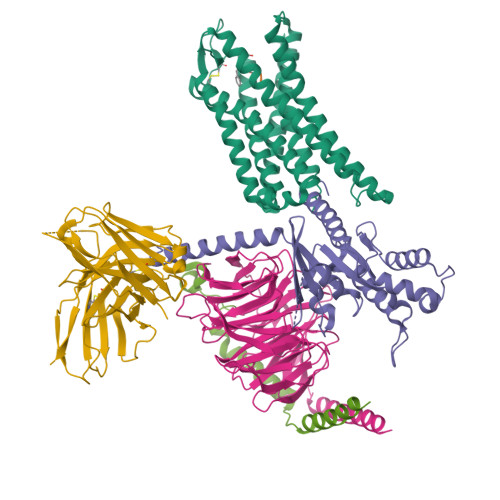
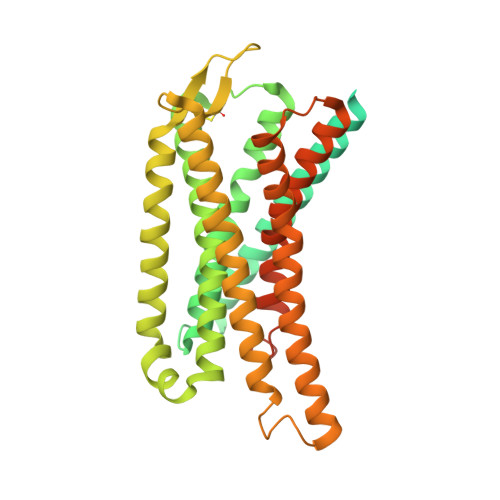
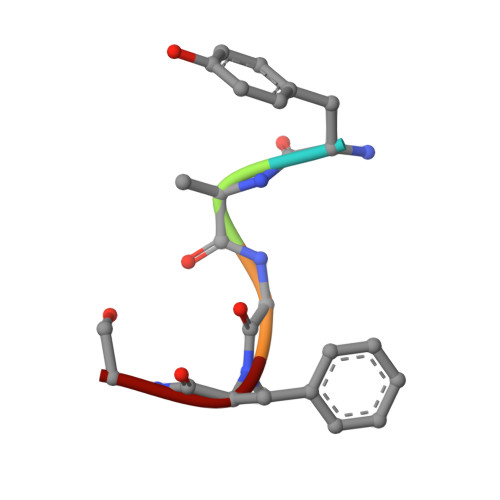
G-protein-coupled receptors (GPCRs) play pivotal roles in various physiological processes. These receptors are activated to different extents by diverse orthosteric ligands and allosteric modulators. However, the mechanisms underlying these variations in signaling activity by allosteric modulators remain largely elusive. Here, we determine the three-dimensional structure of the μ-opioid receptor (MOR), a class A GPCR, in complex with the G i protein and an allosteric modulator, BMS-986122, using cryogenic electron microscopy. Our results reveal that BMS-986122 binding induces changes in the map densities corresponding to R167 3.50 and Y254 5.58 , key residues in the structural motifs conserved among class A GPCRs. Nuclear magnetic resonance analyses of MOR in the absence of the G i protein reveal that BMS-986122 binding enhances the formation of the interaction between R167 3.50 and Y254 5.58 , thus stabilizing the fully-activated conformation, where the intracellular half of TM6 is outward-shifted to allow for interaction with the G i protein. These findings illuminate that allosteric modulators like BMS-986122 can potentiate receptor activation through alterations in the conformational dynamics in the core region of GPCRs. Together, our results demonstrate the regulatory mechanisms of GPCRs, providing insights into the rational development of therapeutics targeting GPCRs.
Organizational Affiliation:
Center for Biosystems Dynamics Research (BDR), RIKEN, Kanagawa, Japan.