Structural insights into transcription activation of the Streptomyces antibiotic regulatory protein, AfsR.
Shi, J., Ye, Z., Feng, Z., Wen, A., Wang, L., Zhang, Z., Xu, L., Song, Q., Wang, F., Liu, T., Wang, S., Feng, Y., Lin, W.(2024) iScience 27: 110421-110421
- PubMed: 39108719
- DOI: https://doi.org/10.1016/j.isci.2024.110421
- Primary Citation of Related Structures:
8K60 - PubMed Abstract:
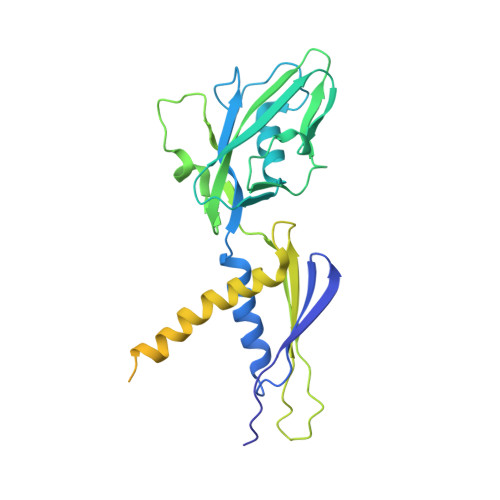
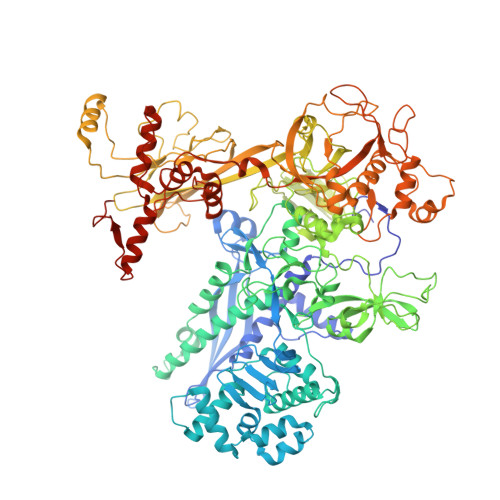
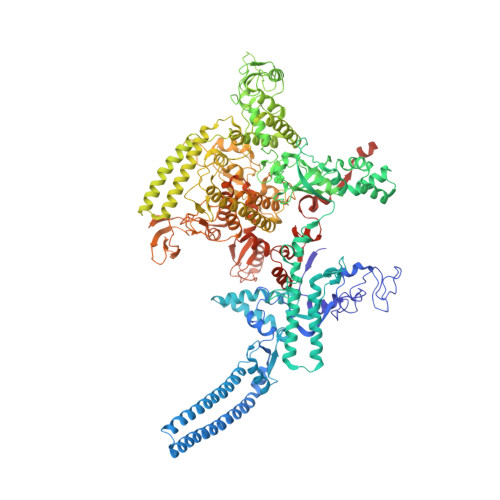
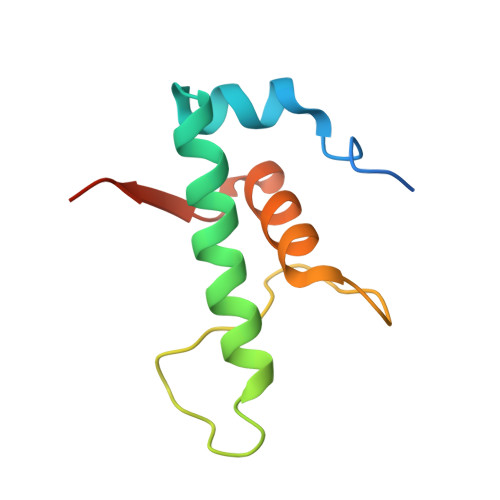
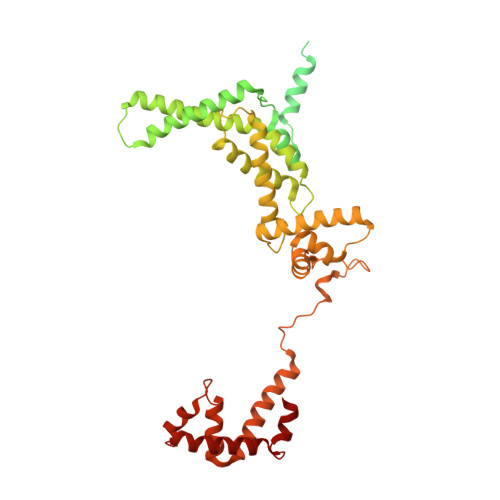
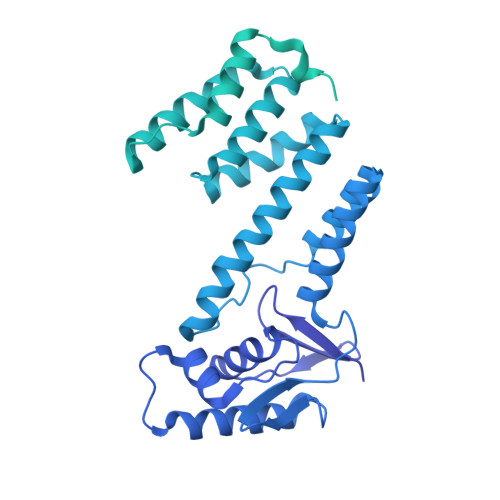
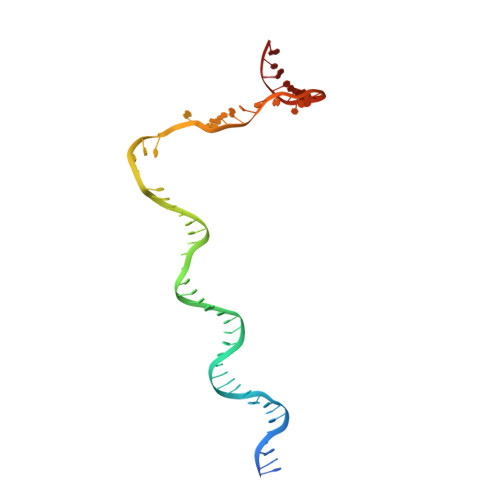
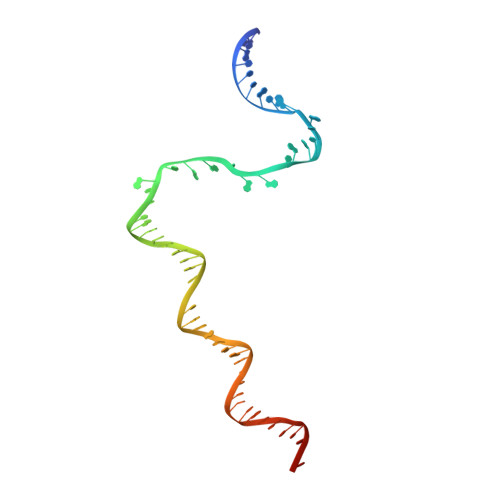
The Streptomyces antibiotic regulatory proteins (SARPs) are ubiquitously distributed transcription activators in Streptomyces and control antibiotics biosynthesis and morphological differentiation. However, the molecular mechanism behind SARP-dependent transcription initiation remains elusive. We here solve the cryo-EM structure of an AfsR-loading RNA polymerase (RNAP)-promoter intermediate complex (AfsR-RPi) including the Streptomyces coelicolor RNAP, a large SARP member AfsR, and its target promoter DNA that retains the upstream portion straight. The structure reveals that one dimeric N-terminal AfsR-SARP domain (AfsR-SARP) specifically engages with the same face of the AfsR-binding sites by the conserved DNA-binding domains (DBDs), replacing σ HrdB R4 to bind the suboptimal -35 element, and shortens the spacer between the -10 and -35 elements. Notably, the AfsR-SARPs also recruit RNAP through extensively interacting with its conserved domains (β flap, σ HrdB R4, and αCTD). Thus, these macromolecular snapshots support a general model and provide valuable clues for SARP-dependent transcription activation in Streptomyces .
- School of Medicine, Nanjing University of Chinese Medicine, Nanjing Drum Tower Hospital, Nanjing 210023, China.
Organizational Affiliation: