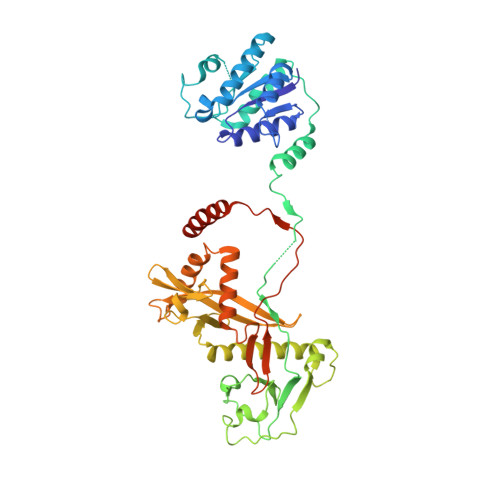
Target ssDNA activates the NADase activity of prokaryotic SPARTA immune system.
Zhang, J.T., Wei, X.Y., Cui, N., Tian, R., Jia, N.(2024) Nat Chem Biol 20: 503-511
- PubMed: 37932528
- DOI: https://doi.org/10.1038/s41589-023-01479-z
- Primary Citation of Related Structures:
8IFK, 8IFL, 8IFM, 8K34 - PubMed Abstract:
Argonaute proteins (Agos), which use small RNAs or DNAs as guides to recognize complementary nucleic acid targets, mediate RNA silencing in eukaryotes. In prokaryotes, Agos are involved in immunity: the short prokaryotic Ago/TIR-APAZ (SPARTA) immune system triggers cell death by degrading NAD + in response to invading plasmids, but its molecular mechanisms remain unknown. Here we used cryo-electron microscopy to determine the structures of inactive monomeric and active tetrameric Crenotalea thermophila SPARTA complexes, revealing mechanisms underlying SPARTA assembly, RNA-guided recognition of target single-stranded DNA (ssDNA) and subsequent SPARTA tetramerization, as well as tetramerization-dependent NADase activation. The small RNA guides Ago to recognize its ssDNA target, inducing SPARTA tetramerization via both Ago- and TIR-mediated interactions and resulting in a two-stranded, parallel, head-to-tail TIR rearrangement primed for NAD + hydrolysis. Our findings thus identify the molecular basis for target ssDNA-mediated SPARTA activation, which will facilitate the development of SPARTA-based biotechnological tools.
Organizational Affiliation:
Department of Biochemistry, School of Medicine, Southern University of Science and Technology, Shenzhen, China.