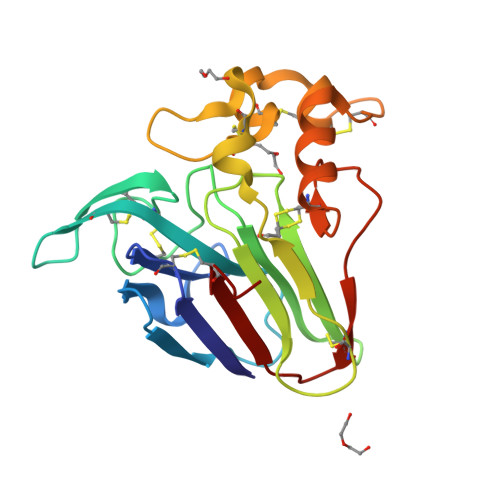
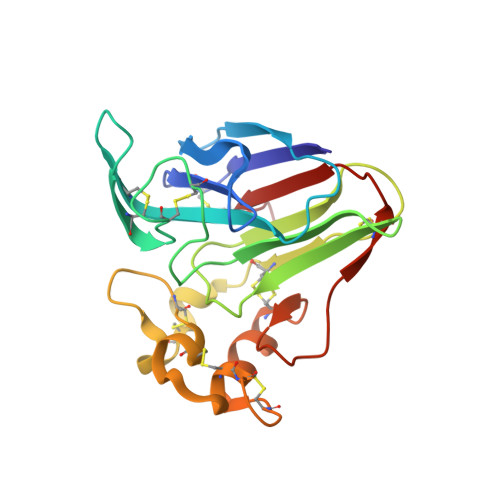
Subatomic structure of orthorhombic thaumatin at 0.89 angstrom reveals that highly flexible conformations are crucial for thaumatin sweetness.
Masuda, T., Suzuki, M., Yamasaki, M., Mikami, B.(2024) Biochem Biophys Res Commun 703: 149601-149601
- PubMed: 38364680
- DOI: https://doi.org/10.1016/j.bbrc.2024.149601
- Primary Citation of Related Structures:
8JZ8 - PubMed Abstract:
Thaumatin is a sweet-tasting protein that elicits a sweet taste at a threshold of approximately 50 nM. Structure-sweetness relationships in thaumatin suggest that the basicity of two amino acids residues, Arg82 and Lys67, are particularly responsible for sweetness. Using tetragonal crystals, our structural analysis suggested that flexible sidechain conformations of these two residues play an important role in sweetness. However, in tetragonal crystals, Arg82 is adjacent to symmetry-related residues, and its flexibility is relatively restrained by the crystal packing. To reduce and diminish these symmetry-related effects, orthorhombic crystals were prepared, and their structures were successfully determined at a resolution of 0.89 Å. Within the orthorhombic lattice, two alternative conformations were more clearly visible at Lys67 than in a tetragonal system. Interestingly, for the first time, three alternative conformations at Arg82 were only found in an orthorhombic system. These results suggest the importance of flexible conformations in sweetness determinants. Such subtle structural variations might serve to adjust the complementarity of the electrostatic potentials of sweet receptors, thereby eliciting the potent sweet taste of thaumatin.
Organizational Affiliation:
Department of Food Sciences and Human Nutrition, Faculty of Agriculture, Ryukoku University, 1-5 Yokotani, Seta Oe-cho, Otsu, Shiga, 520-2194, Japan; Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Gokasho, Uji, Kyoto, 611-0011, Japan. Electronic address: t2masuda@agr.ryukoku.ac.jp.