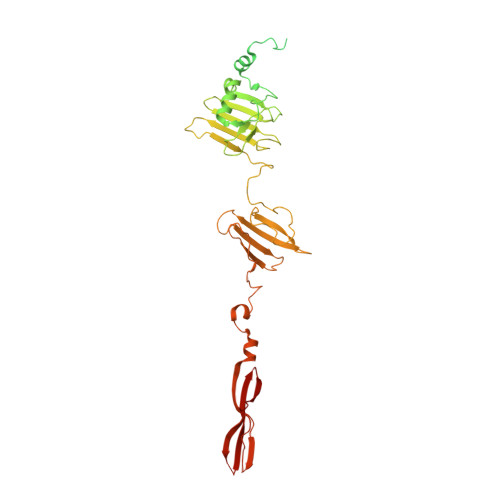
Determination of the three-dimensional structure of bacteriophage Mu(-) tail fiber and its characterization.
Mori, Y., Yamashita, E., Nakagawa, A., Matsuzawa, T., Inagaki, M., Aiba, Y., Tanaka, S., Hatori, S., Ayami, M., Takeda, S.(2024) Virology 593: 110017-110017
- PubMed: 38382161
- DOI: https://doi.org/10.1016/j.virol.2024.110017
- Primary Citation of Related Structures:
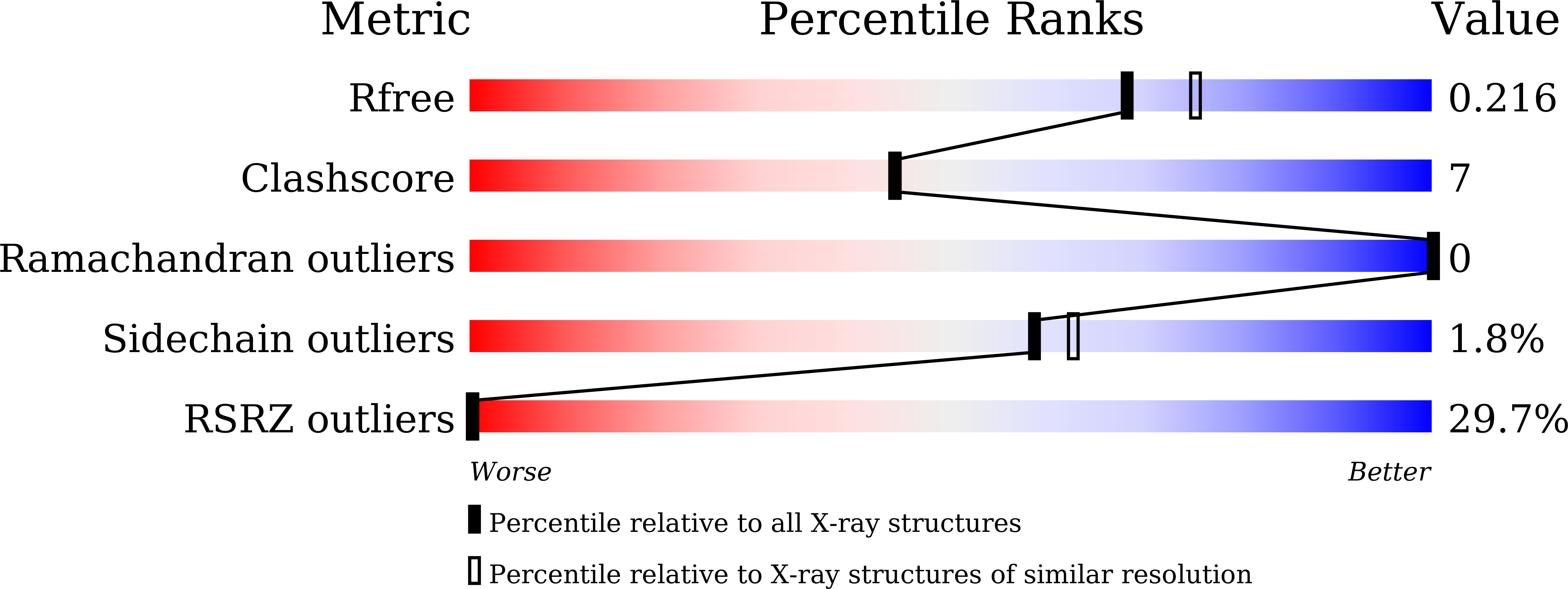
8JU3 - PubMed Abstract:
Bacteriophage Mu is a temperate phage known to infect various species of Enterobacteria, playing a role in bacterial mutation induction and horizontal gene transfer. The phage possesses two types of tail fibers important for host recognition, which enable it to expand its range of hosts. The alternate tail fibers are formed through the action of genes 49-50 or 52-51, allowing the Mu phage to recognize different surfaces of host cells. In a previous study, we presented the X-ray crystal structure of the C-terminal lipopolysaccharide (LPS)-binding domain of gene product (gp) 49, one of the subunits comprising the Mu tail fiber. In this study, we have determined the structure of the alternative tail fiber subunit, gp52, and compared it with other tail fibers. The results revealed that Mu phage employs different structural motifs for two individual tail fibers for recognizing different hosts.
Organizational Affiliation:
Faculty of Science and Technology, Division of Molecular Science, Gunma University, 1-5-1 Tenjin-cho, Kiryu, Gunma, 376-8515, Japan.