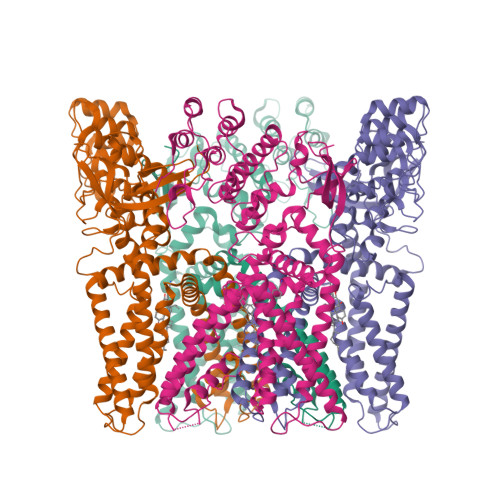
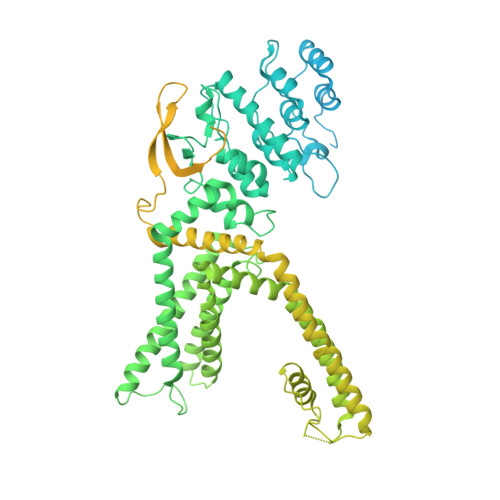
Structural basis of TRPV1 inhibition by SAF312 and cholesterol.
Fan, J., Ke, H., Lei, J., Wang, J., Tominaga, M., Lei, X.(2024) Nat Commun 15: 6689-6689
- PubMed: 39107321
- DOI: https://doi.org/10.1038/s41467-024-51085-3
- Primary Citation of Related Structures:
8JQR, 8X94 - PubMed Abstract:
Transient Receptor Potential Vanilloid 1 (TRPV1) plays a central role in pain sensation and is thus an attractive pharmacological drug target. SAF312 is a potent, selective, and non-competitive antagonist of TRPV1 and shows promising potential in treating ocular surface pain. However, the precise mechanism by which SAF312 inhibits TRPV1 remains poorly understood. Here, we present the cryo-EM structure of human TRPV1 in complex with SAF312, elucidating the structural foundation of its antagonistic effects on TRPV1. SAF312 binds to the vanilloid binding pocket, preventing conformational changes in S4 and S5 helices, which are essential for channel gating. Unexpectedly, a putative cholesterol was found to contribute to SAF312's inhibition. Complemented by mutagenesis experiments and molecular dynamics simulations, our research offers substantial mechanistic insights into the regulation of TRPV1 by SAF312, highlighting the interplay between the antagonist and cholesterol in modulating TRPV1 function. This work not only expands our understanding of TRPV1 inhibition by SAF312 but also lays the groundwork for further developments in the design and optimization of TRPV1-related therapies.
Organizational Affiliation:
Beijing National Laboratory for Molecular Sciences, Key Laboratory of Bioorganic Chemistry and Molecular Engineering of Ministry of Education, Institute of Organic Chemistry, College of Chemistry and Molecular Engineering, Peking University, Beijing, 100871, China. fanjp@pku.edu.cn.