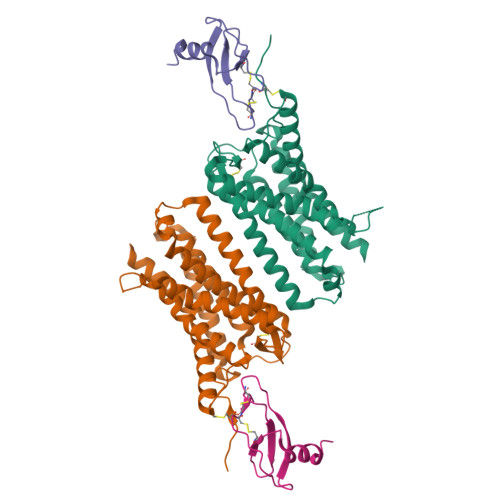
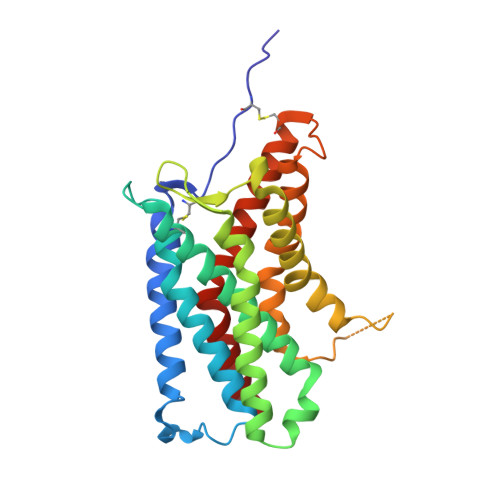
Molecular mechanism of distinct chemokine engagement and functional divergence of the human Duffy antigen receptor.
Saha, S., Khanppnavar, B., Maharana, J., Kim, H., Carino, C.M.C., Daly, C., Houston, S., Sharma, S., Zaidi, N., Dalal, A., Mishra, S., Ganguly, M., Tiwari, D., Kumari, P., Jhingan, G.D., Yadav, P.N., Plouffe, B., Inoue, A., Chung, K.Y., Banerjee, R., Korkhov, V.M., Shukla, A.K.(2024) Cell 187: 4751-4769.e25
- PubMed: 39089252
- DOI: https://doi.org/10.1016/j.cell.2024.07.005
- Primary Citation of Related Structures:
8JPS - PubMed Abstract:
The Duffy antigen receptor is a seven-transmembrane (7TM) protein expressed primarily at the surface of red blood cells and displays strikingly promiscuous binding to multiple inflammatory and homeostatic chemokines. It serves as the basis of the Duffy blood group system in humans and also acts as the primary attachment site for malarial parasite Plasmodium vivax and pore-forming toxins secreted by Staphylococcus aureus. Here, we comprehensively profile transducer coupling of this receptor, discover potential non-canonical signaling pathways, and determine the cryoelectron microscopy (cryo-EM) structure in complex with the chemokine CCL7. The structure reveals a distinct binding mode of chemokines, as reflected by relatively superficial binding and a partially formed orthosteric binding pocket. We also observe a dramatic shortening of TM5 and 6 on the intracellular side, which precludes the formation of the docking site for canonical signal transducers, thereby providing a possible explanation for the distinct pharmacological and functional phenotype of this receptor.
Organizational Affiliation:
Department of Biological Sciences and Bioengineering, Indian Institute of Technology Kanpur, Kanpur 208016, India.