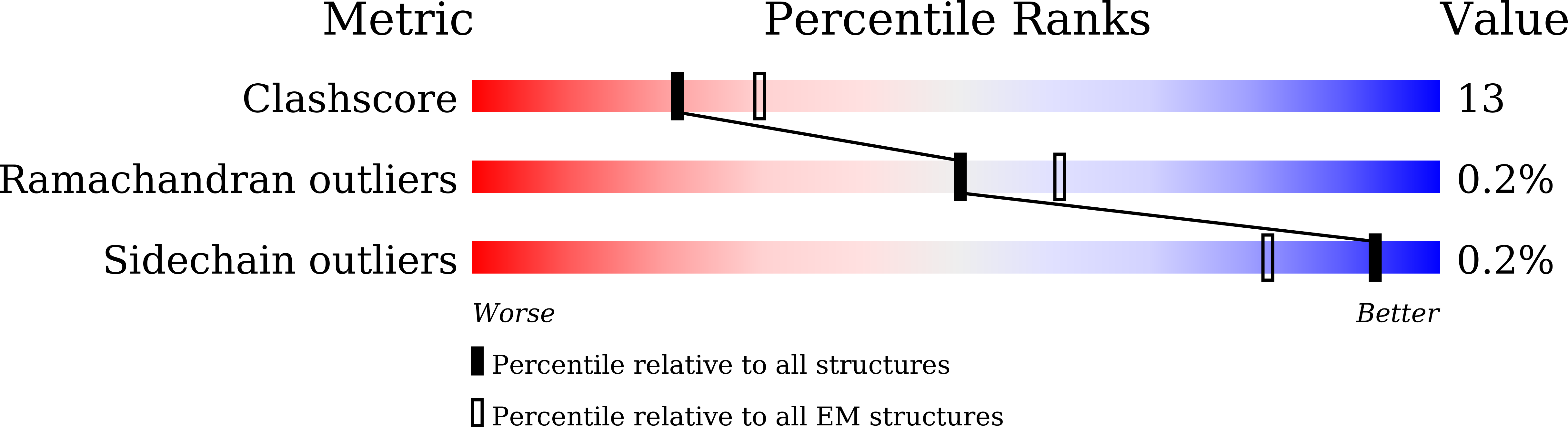Structural basis for antiepileptic drugs and botulinum neurotoxin recognition of SV2A.
Yamagata, A., Ito, K., Suzuki, T., Dohmae, N., Terada, T., Shirouzu, M.(2024) Nat Commun 15: 3027-3027
- PubMed: 38637505
- DOI: https://doi.org/10.1038/s41467-024-47322-4
- Primary Citation of Related Structures:
8JLC, 8JLE, 8JLF, 8JLG, 8JLH, 8JLI, 8JS8, 8JS9, 8K77 - PubMed Abstract:
More than one percent of people have epilepsy worldwide. Levetiracetam (LEV) is a successful new-generation antiepileptic drug (AED), and its derivative, brivaracetam (BRV), shows improved efficacy. Synaptic vesicle glycoprotein 2a (SV2A), a putative membrane transporter in the synaptic vesicles (SVs), has been identified as a target of LEV and BRV. SV2A also serves as a receptor for botulinum neurotoxin (BoNT), which is the most toxic protein and has paradoxically emerged as a potent reagent for therapeutic and cosmetic applications. Nevertheless, no structural analysis on AEDs and BoNT recognition by full-length SV2A has been available. Here we describe the cryo-electron microscopy structures of the full-length SV2A in complex with the BoNT receptor-binding domain, BoNT/A2 H C, and either LEV or BRV. The large fourth luminal domain of SV2A binds to BoNT/A2 H C through protein-protein and protein-glycan interactions. LEV and BRV occupy the putative substrate-binding site in an outward-open conformation. A propyl group in BRV creates additional contacts with SV2A, explaining its higher binding affinity than that of LEV, which was further supported by label-free spectral shift assay. Numerous LEV derivatives have been developed as AEDs and positron emission tomography (PET) tracers for neuroimaging. Our work provides a structural framework for AEDs and BoNT recognition of SV2A and a blueprint for the rational design of additional AEDs and PET tracers.
Organizational Affiliation:
Laboratory for Protein Functional and Structural Biology, RIKEN Center for Biosystems Dynamics Research, 1-7-22 Suehiro-cho, Tsurumi-ku, Yokohama, Kanagawa, Japan. atsushi.yamagata@riken.jp.