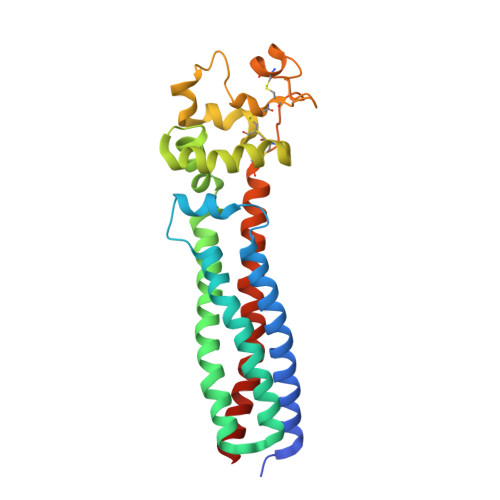
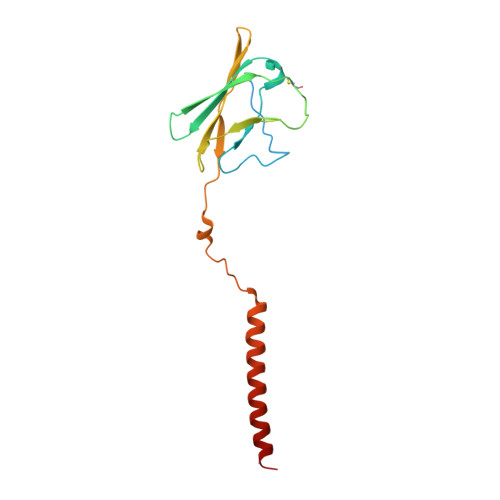
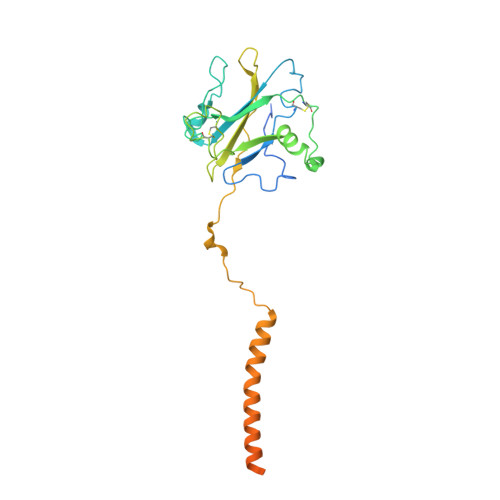
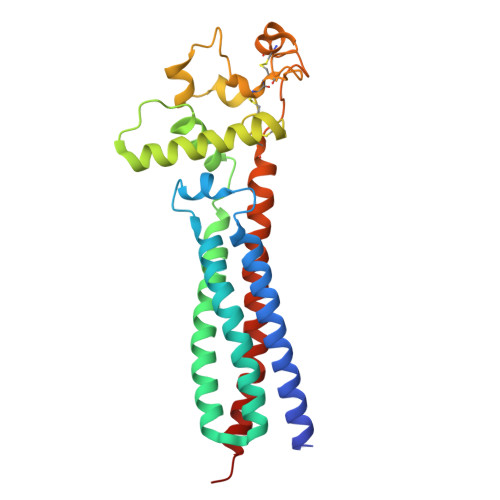
Cryo-EM elucidates the uroplakin complex structure within liquid-crystalline lipids in the porcine urothelial membrane.
Yanagisawa, H., Kita, Y., Oda, T., Kikkawa, M.(2023) Commun Biol 6: 1018-1018
- PubMed: 37805589
- DOI: https://doi.org/10.1038/s42003-023-05393-x
- Primary Citation of Related Structures:
8JJ5 - PubMed Abstract:
The urothelium, a distinct epithelial tissue lining the urinary tract, serves as an essential component in preserving urinary tract integrity and thwarting infections. The asymmetric unit membrane (AUM), primarily composed of the uroplakin complex, constitutes a critical permeability barrier in fulfilling this role. However, the molecular architectures of both the AUM and the uroplakin complex have remained enigmatic due to the paucity of high-resolution structural data. In this study, we utilized cryo-electron microscopy to elucidate the three-dimensional structure of the uroplakin complex within the porcine AUM. While the global resolution achieved was 3.5 Å, we acknowledge that due to orientation bias, the resolution in the vertical direction was determined to be 6.3 Å. Our findings unveiled that the uroplakin complexes are situated within hexagonally arranged crystalline lipid membrane domains, rich in hexosylceramides. Moreover, our research rectifies a misconception in a previous model by confirming the existence of a domain initially believed to be absent, and pinpointing the accurate location of a crucial Escherichia coli binding site implicated in urinary tract infections. These discoveries offer valuable insights into the molecular underpinnings governing the permeability barrier function of the urothelium and the orchestrated lipid phase formation within the plasma membrane.
Organizational Affiliation:
Department of Cell Biology and Anatomy, Graduate School of Medicine, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo, 113-0033, Japan.