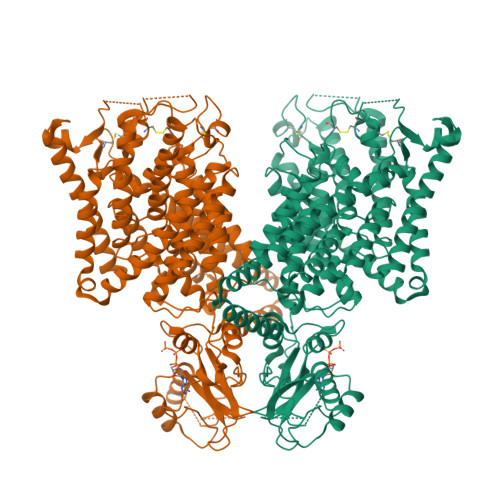
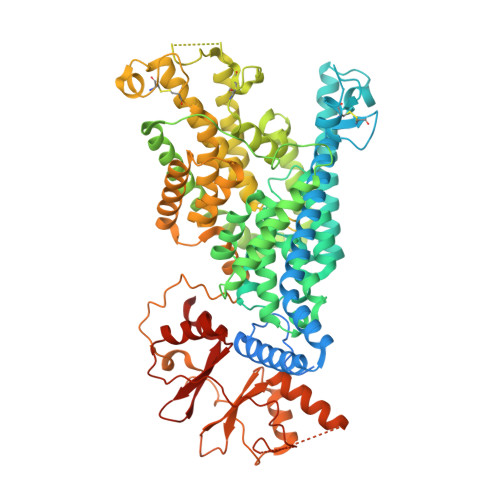
Structural basis of adenine nucleotides regulation and neurodegenerative pathology in ClC-3 exchanger.
Wan, Y., Guo, S., Zhen, W., Xu, L., Chen, X., Liu, F., Shen, Y., Liu, S., Hu, L., Wang, X., Ye, F., Wang, Q., Wen, H., Yang, F.(2024) Nat Commun 15: 6654-6654
- PubMed: 39107281
- DOI: https://doi.org/10.1038/s41467-024-50975-w
- Primary Citation of Related Structures:
8JEV, 8JGJ, 8JGK, 8JGL, 8JGS, 8JGV - PubMed Abstract:
The ClC-3 chloride/proton exchanger is both physiologically and pathologically critical, as it is potentiated by ATP to detect metabolic energy level and point mutations in ClC-3 lead to severe neurodegenerative diseases in human. However, why this exchanger is differentially modulated by ATP, ADP or AMP and how mutations caused gain-of-function remains largely unknow. Here we determine the high-resolution structures of dimeric wildtype ClC-3 in the apo state and in complex with ATP, ADP and AMP, and the disease-causing I607T mutant in the apo and ATP-bounded state by cryo-electron microscopy. In combination with patch-clamp recordings and molecular dynamic simulations, we reveal how the adenine nucleotides binds to ClC-3 and changes in ion occupancy between apo and ATP-bounded state. We further observe I607T mutation induced conformational changes and augments in current. Therefore, our study not only lays the structural basis of adenine nucleotides regulation in ClC-3, but also clearly indicates the target region for drug discovery against ClC-3 mediated neurodegenerative diseases.
Organizational Affiliation:
Department of Biophysics and Disease Center of the First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang, China.