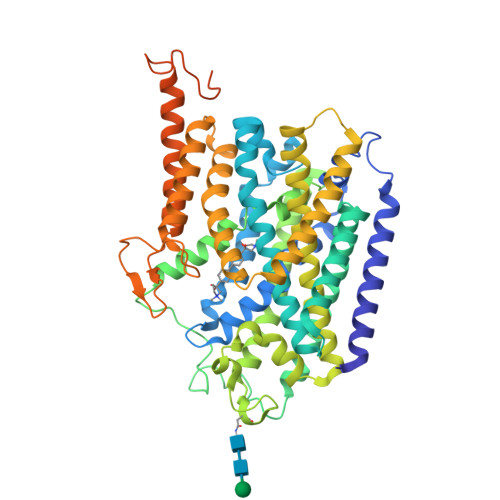
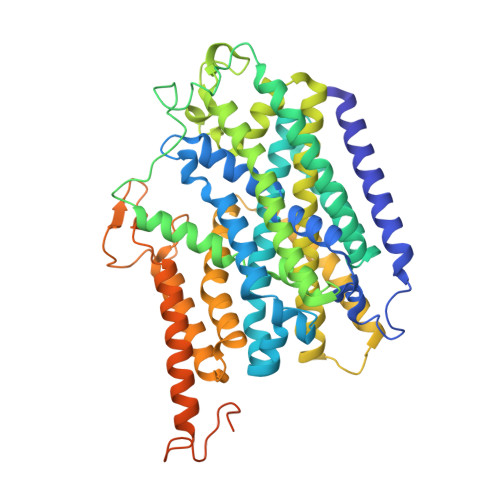
Transport mechanism of presynaptic high-affinity choline uptake by CHT1.
Qiu, Y., Gao, Y., Huang, B., Bai, Q., Zhao, Y.(2024) Nat Struct Mol Biol 31: 701-709
- PubMed: 38589607
- DOI: https://doi.org/10.1038/s41594-024-01259-w
- Primary Citation of Related Structures:
8J74, 8J75, 8J76, 8J77 - PubMed Abstract:
Choline is a vital nutrient and a precursor for the biosynthesis of essential metabolites, including acetylcholine (ACh), that play a central role in fetal development, especially in the brain. In cholinergic neurons, the high-affinity choline transporter (CHT1) provides an extraordinarily efficient reuptake mechanism to reutilize choline derived from intrasynaptical ACh hydrolysis and maintain ACh synthesis in the presynapse. Here, we determined structures of human CHT1 in three discrete states: the outward-facing state bound with the competitive inhibitor hemicholinium-3 (HC-3); the inward-facing occluded state bound with the substrate choline; and the inward-facing apo open state. Our structures and functional characterizations elucidate how the inhibitor and substrate are recognized. Moreover, our findings shed light on conformational changes when transitioning from an outward-facing to an inward-facing state and establish a framework for understanding the transport cycle, which relies on the stabilization of the outward-facing state by a short intracellular helix, IH1.
Organizational Affiliation:
Key Laboratory of Biomacromolecules (CAS), National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, China.