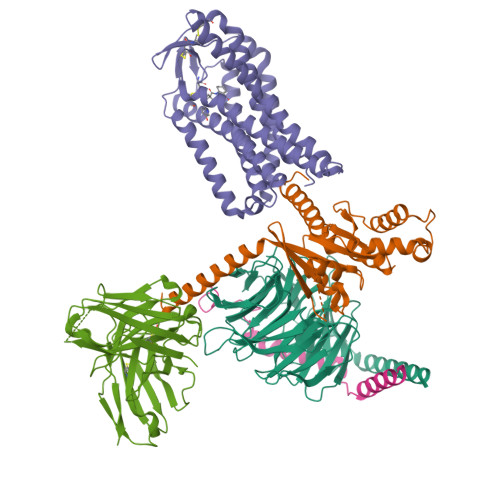
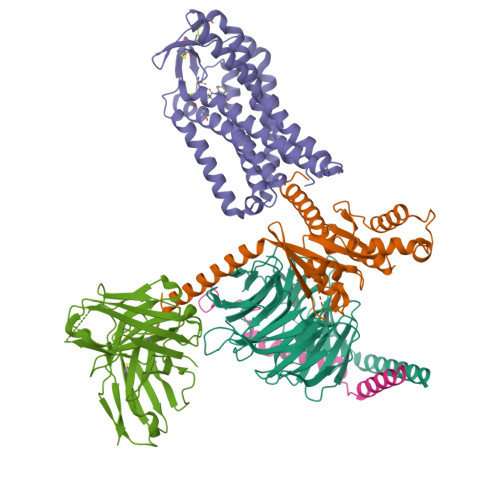
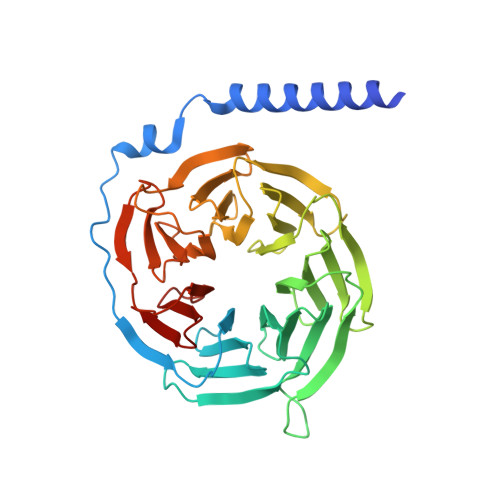
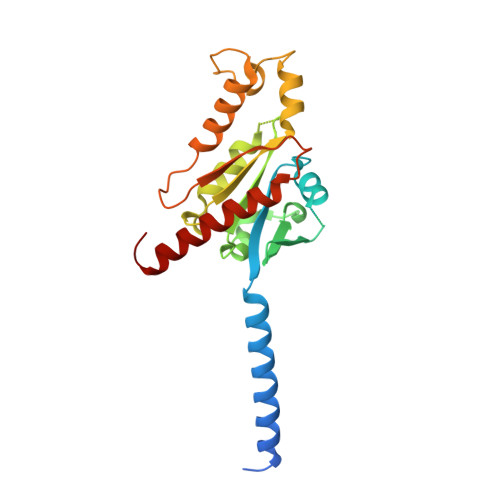
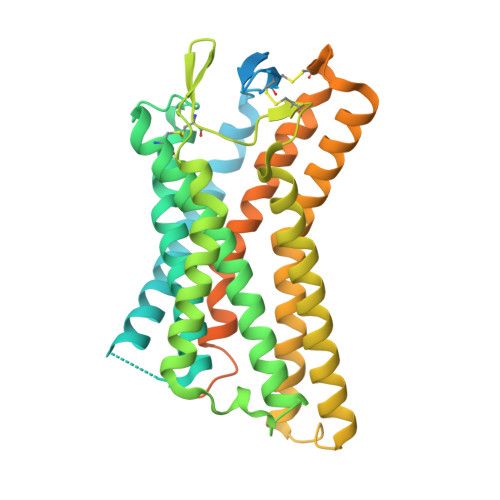
Structure-guided engineering of biased-agonism in the human niacin receptor via single amino acid substitution.
Yadav, M.K., Sarma, P., Maharana, J., Ganguly, M., Mishra, S., Zaidi, N., Dalal, A., Singh, V., Saha, S., Mahajan, G., Sharma, S., Chami, M., Banerjee, R., Shukla, A.K.(2024) Nat Commun 15: 1939-1939
- PubMed: 38431681
- DOI: https://doi.org/10.1038/s41467-024-46239-2
- Primary Citation of Related Structures:
8IY9, 8IYH, 8IYW, 8JER, 8JHN - PubMed Abstract:
The Hydroxycarboxylic acid receptor 2 (HCA2), also known as the niacin receptor or GPR109A, is a prototypical GPCR that plays a central role in the inhibition of lipolytic and atherogenic activities. Its activation also results in vasodilation that is linked to the side-effect of flushing associated with dyslipidemia drugs such as niacin. GPR109A continues to be a target for developing potential therapeutics in dyslipidemia with minimized flushing response. Here, we present cryo-EM structures of the GPR109A in complex with dyslipidemia drugs, niacin or acipimox, non-flushing agonists, MK6892 or GSK256073, and recently approved psoriasis drug, monomethyl fumarate (MMF). These structures elucidate the binding mechanism of agonists, molecular basis of receptor activation, and insights into biased signaling elicited by some of the agonists. The structural framework also allows us to engineer receptor mutants that exhibit G-protein signaling bias, and therefore, our study may help in structure-guided drug discovery efforts targeting this receptor.
Organizational Affiliation:
Department of Biological Sciences and Bioengineering, Indian Institute of Technology, Kanpur, 08016, India.