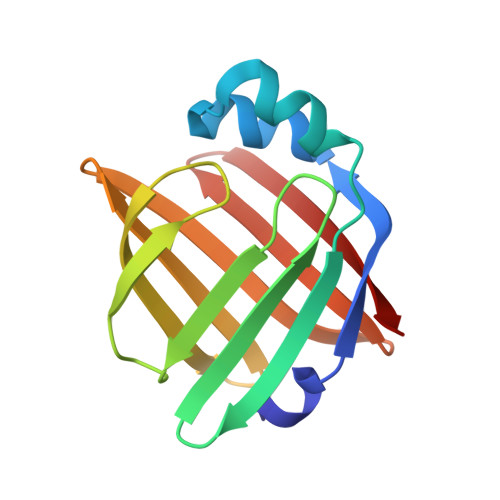
Fatty acid-binding proteins 3, 7, and 8 bind cholesterol and facilitate its egress from lysosomes.
Fang, X.X., Wei, P., Zhao, K., Sheng, Z.C., Song, B.L., Yin, L., Luo, J.(2024) J Cell Biol 223
- PubMed: 38429999
- DOI: https://doi.org/10.1083/jcb.202211062
- Primary Citation of Related Structures:
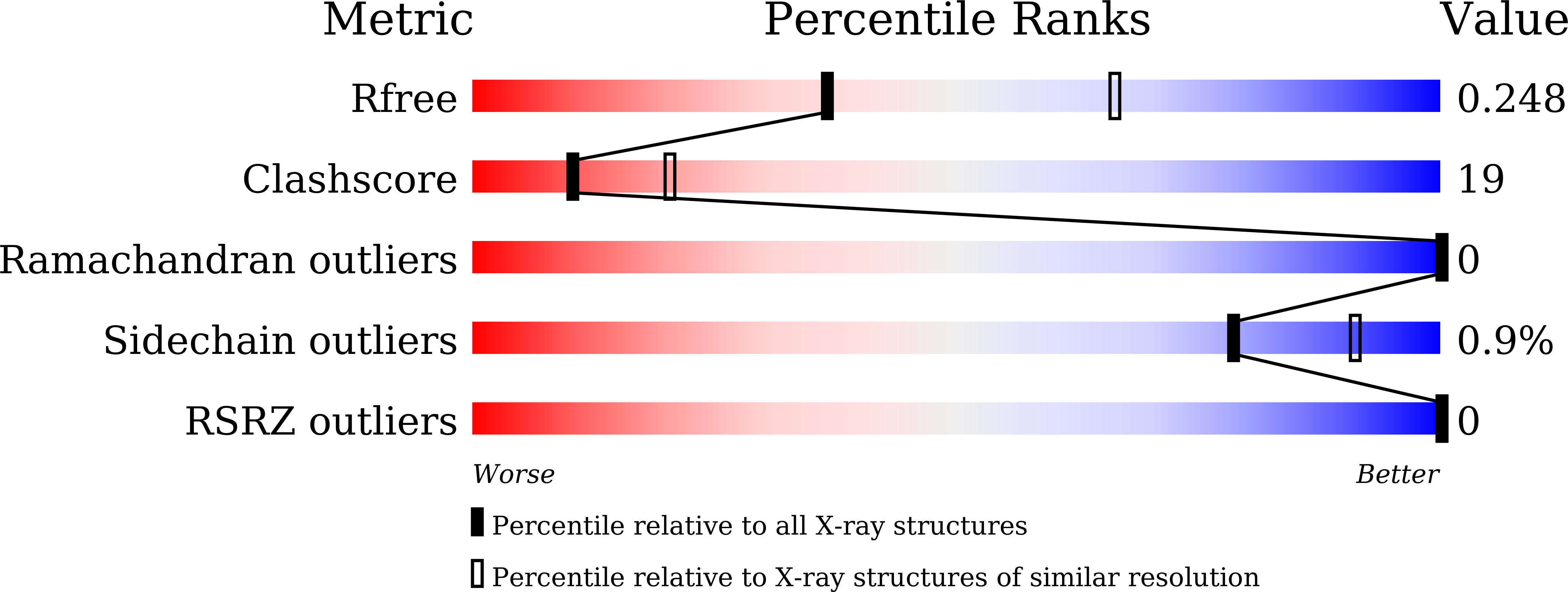
8IVF, 8IVL - PubMed Abstract:
Cholesterol from low-density lipoprotein (LDL) can be transported to many organelle membranes by non-vesicular mechanisms involving sterol transfer proteins (STPs). Fatty acid-binding protein (FABP) 7 was identified in our previous study searching for new regulators of intracellular cholesterol trafficking. Whether FABP7 is a bona fide STP remains unknown. Here, we found that FABP7 deficiency resulted in the accumulation of LDL-derived cholesterol in lysosomes and reduced cholesterol levels on the plasma membrane. A crystal structure of human FABP7 protein in complex with cholesterol was resolved at 2.7 Å resolution. In vitro, FABP7 efficiently transported the cholesterol analog dehydroergosterol between the liposomes. Further, the silencing of FABP3 and 8, which belong to the same family as FABP7, caused robust cholesterol accumulation in lysosomes. These two FABP proteins could transport dehydroergosterol in vitro as well. Collectively, our results suggest that FABP3, 7, and 8 are a new class of STPs mediating cholesterol egress from lysosomes.
Organizational Affiliation:
The Institute for Advanced Studies, College of Life Sciences, Hubei Key Laboratory of Cell Homeostasis, Taikang Center for Life and Medical Sciences, Taikang Medical School, Frontier Science Center for Immunology and Metabolism, Wuhan University, Wuhan, China.