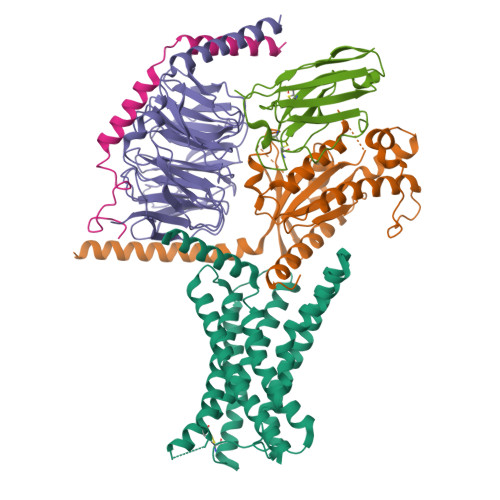
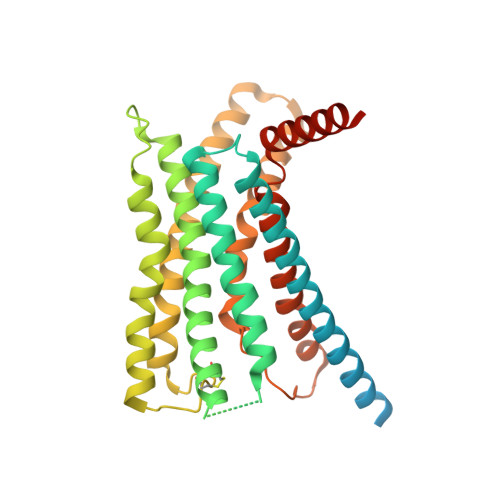
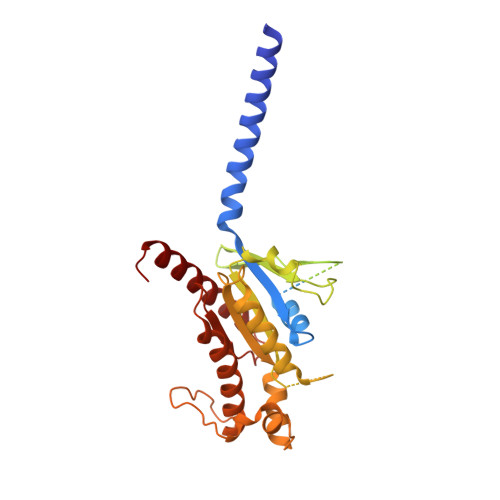
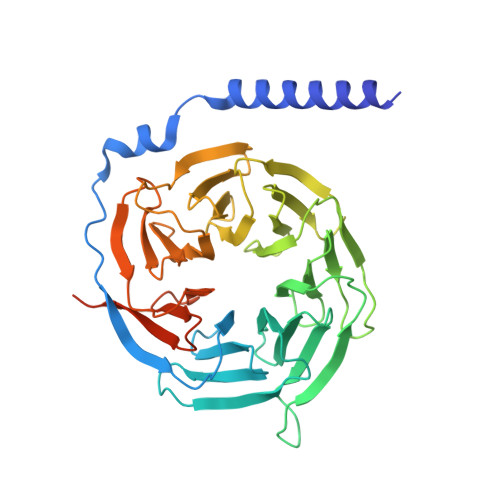
Molecular basis of signal transduction mediated by the human GIPR splice variants.
Zhao, F., Hang, K., Zhou, Q., Shao, L., Li, H., Li, W., Lin, S., Dai, A., Cai, X., Liu, Y., Xu, Y., Feng, W., Yang, D., Wang, M.W.(2023) Proc Natl Acad Sci U S A 120: e2306145120-e2306145120
- PubMed: 37792509
- DOI: https://doi.org/10.1073/pnas.2306145120
- Primary Citation of Related Structures:
8ITL, 8ITM - PubMed Abstract:
Glucose-dependent insulinotropic polypeptide receptor (GIPR) is a potential drug target for metabolic disorders. It works with glucagon-like peptide-1 receptor and glucagon receptor in humans to maintain glucose homeostasis. Unlike the other two receptors, GIPR has at least 13 reported splice variants (SVs), more than half of which have sequence variations at either C or N terminus. To explore their roles in endogenous peptide-mediated GIPR signaling, we determined the cryoelectron microscopy (cryo-EM) structures of the two N terminus-altered SVs (referred as GIPR-202 and GIPR-209 in the Ensembl database, SV1 and SV2 here, respectively) and investigated the outcome of coexpressing each of them in question with GIPR in HEK293T cells with respect to ligand binding, receptor expression, cAMP (adenosine 3,5-cyclic monophosphate) accumulation, β-arrestin recruitment, and cell surface localization. It was found that while both N terminus-altered SVs of GIPR neither bound to the hormone nor elicited signal transduction per se, they suppressed ligand binding and cAMP accumulation of GIPR. Meanwhile, SV1 reduced GIPR-mediated β-arrestin 2 responses. The cryo-EM structures of SV1 and SV2 showed that they reorganized the extracellular halves of transmembrane helices 1, 6, and 7 and extracellular loops 2 and 3 to adopt a ligand-binding pocket-occupied conformation, thereby losing binding ability to the peptide. The results suggest a form of signal bias that is constitutive and ligand-independent, thus expanding our knowledge of biased signaling beyond pharmacological manipulation (i.e., ligand specific) as well as constitutive and ligand-independent (e.g., SV1 of the growth hormone-releasing hormone receptor).
Organizational Affiliation:
The National Center for Drug Screening, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China.