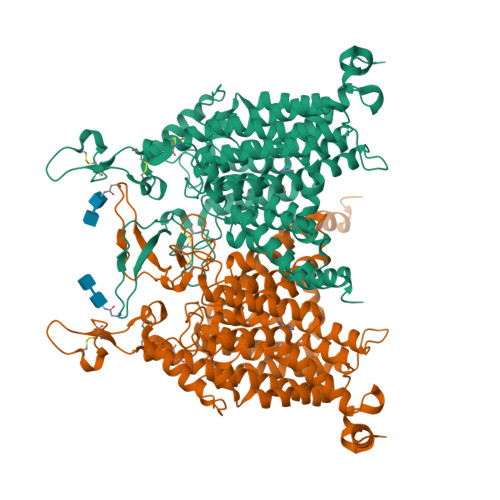
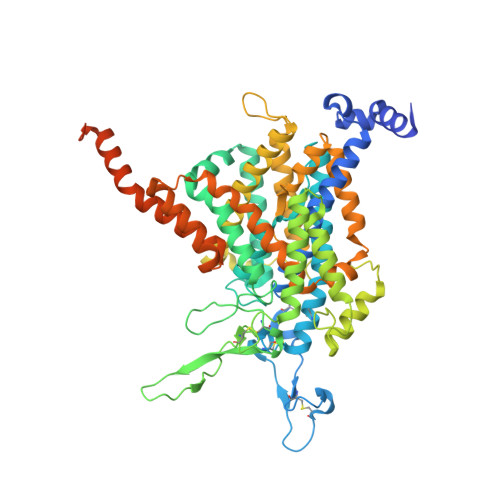
Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Xu, L., Jia, W., Tao, X., Ye, F., Zhang, Y., Ding, Z.J., Zheng, S.J., Qiao, S., Su, N., Zhang, Y., Wu, S., Guo, J.(2024) Nat Plants 10: 180-191
- PubMed: 38172575
- DOI: https://doi.org/10.1038/s41477-023-01590-y
- Primary Citation of Related Structures:
8IRL, 8IRM, 8IRN, 8IRO, 8IRP, 8WMQ, 8WO7 - PubMed Abstract:
Cytokinins are essential for plant growth and development, and their tissue distributions are regulated by transmembrane transport. Recent studies have revealed that members of the 'Aza-Guanine Resistant' (AZG) protein family from Arabidopsis thaliana can mediate cytokinin uptake in roots. Here we present 2.7 to 3.3 Å cryo-electron microscopy structures of Arabidopsis AZG1 in the apo state and in complex with its substrates trans-zeatin (tZ), 6-benzyleaminopurine (6-BAP) or kinetin. AZG1 forms a homodimer and each subunit shares a similar topology and domain arrangement with the proteins of the nucleobase/ascorbate transporter (NAT) family. These structures, along with functional analyses, reveal the molecular basis for cytokinin recognition. Comparison of the AZG1 structures determined in inward-facing conformations and predicted by AlphaFold2 in the occluded conformation allowed us to propose that AZG1 may carry cytokinins across the membrane through an elevator mechanism.
Organizational Affiliation:
Department of Biophysics and Department of Neurology of the Fourth Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang, China. lingyixv@zju.edu.cn.