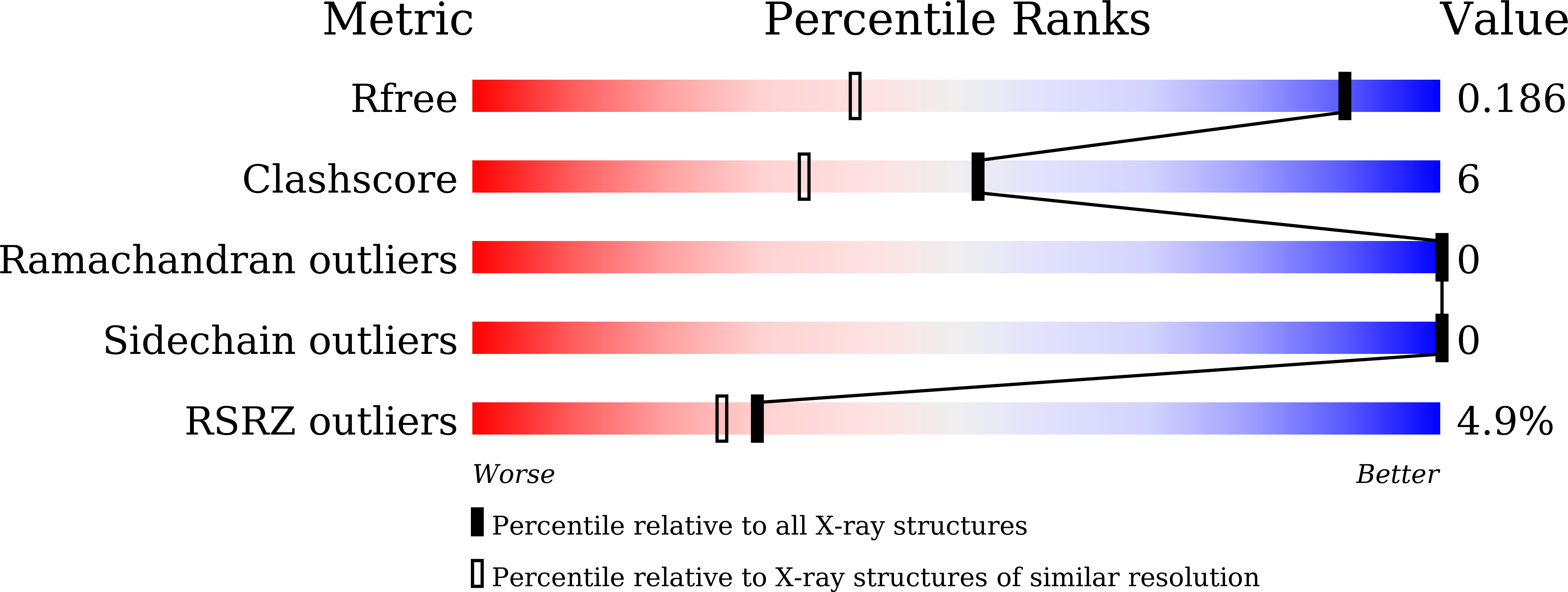
Polymorphic nanobody crystals as long-acting intravitreal therapy for wet age-related macular degeneration.
Zhu, S., Fan, S., Tang, T., Huang, J., Zhou, H., Huang, C., Chen, Y., Qian, F.(2023) Bioeng Transl Med 8: e10523-e10523
- PubMed: 38023710
- DOI: https://doi.org/10.1002/btm2.10523
- Primary Citation of Related Structures:
8IIU, 8IJS, 8IJZ - PubMed Abstract:
Wet age-related macular degeneration (wet AMD) is the most common cause of blindness, and chronic intravitreal injection of anti-vascular endothelial growth factor (VEGF) proteins has been the dominant therapeutic approach. Less intravitreal injection and a prolonged inter-injection interval are the main drivers behind new wet AMD drug innovations. By rationally engineering the surface residues of a model anti-VEGF nanobody, we obtained a series of anti-VEGF nanobodies with identical protein structures and VEGF binding affinities, while drastically different crystallization propensities and crystal lattice structures. Among these nanobody crystals, the P 2 1 2 1 2 1 lattice appeared to be denser and released protein slower than the P 1 lattice, while nanobody crystals embedding zinc coordination further slowed the protein release rate. The polymorphic protein crystals could be a potentially breakthrough strategy for chronic intravitreal administration of anti-VEGF proteins.
Organizational Affiliation:
School of Pharmaceutical Sciences, Beijing Advanced Innovation Center for Structural Biology, and Key Laboratory of Bioorganic Phosphorus Chemistry and Chemical Biology (Ministry of Education) Tsinghua University Beijing People's Republic of China.