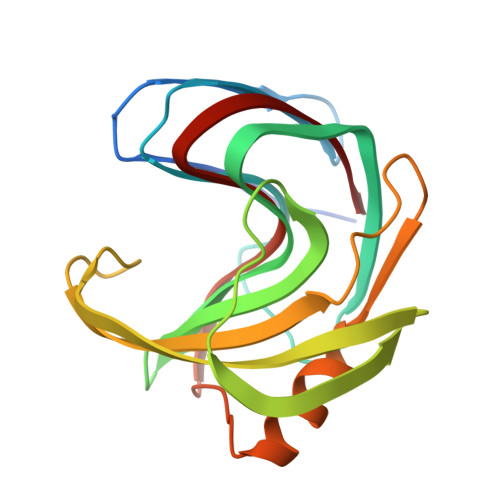
Characterization and structural analysis of the endo-1,4-beta-xylanase GH11 from the hemicellulose-degrading Thermoanaerobacterium saccharolyticum useful for lignocellulose saccharification.
Kim, I.J., Kim, S.R., Kim, K.H., Bornscheuer, U.T., Nam, K.H.(2023) Sci Rep 13: 17332-17332
- PubMed: 37833340
- DOI: https://doi.org/10.1038/s41598-023-44495-8
- Primary Citation of Related Structures:
8IH0, 8IH1 - PubMed Abstract:
Xylanases are important for the enzymatic breakdown of lignocellulose-based biomass to produce biofuels and other value-added products. We report functional and structural analyses of TsaGH11, an endo-1,4-β-xylanase from the hemicellulose-degrading bacterium, Thermoanaerobacterium saccharolyticum. TsaGH11 was shown to be a thermophilic enzyme that favors acidic conditions with maximum activity at pH 5.0 and 70 °C. It decomposes xylans from beechwood and oat spelts to xylose-containing oligosaccharides with specific activities of 5622.0 and 3959.3 U mg -1 , respectively. The kinetic parameters, K m and k cat towards beechwood xylan, are 12.9 mg mL -1 and 34,015.3 s -1 , respectively, resulting in k cat /K m value of 2658.7 mL mg -1 s -1 , higher by 10 2 -10 3 orders of magnitude compared to other reported GH11s investigated with the same substrate, demonstrating its superior catalytic performance. Crystal structures of TsaGH11 revealed a β-jelly roll fold, exhibiting open and close conformations of the substrate-binding site by distinct conformational flexibility to the thumb region of TsaGH11. In the room-temperature structure of TsaGH11 determined by serial synchrotron crystallography, the electron density map of the thumb domain of the TsaGH11 molecule, which does not affect crystal packing, is disordered, indicating that the thumb domain of TsaGH11 has high structural flexibility at room temperature, with the water molecules in the substrate-binding cleft being more disordered than those in the cryogenic structure. These results expand our knowledge of GH11 structural flexibility at room temperature and pave the way for its application in industrial biomass degradation.
- Department of Food Science and Technology, Institute of Agriculture and Life Science, Gyeongsang National University, Jinju, 52828, South Korea.
Organizational Affiliation: