Crystal structure of reducing-end xylose-releasing exoxylanase in subfamily 7 of glycoside hydrolase family 30.
Nakamichi, Y., Watanabe, M., Fujii, T., Inoue, H., Morita, T.(2023) Proteins 91: 1341-1350
- PubMed: 37144255
- DOI: https://doi.org/10.1002/prot.26505
- Primary Citation of Related Structures:
8IDP, 8IDQ - PubMed Abstract:
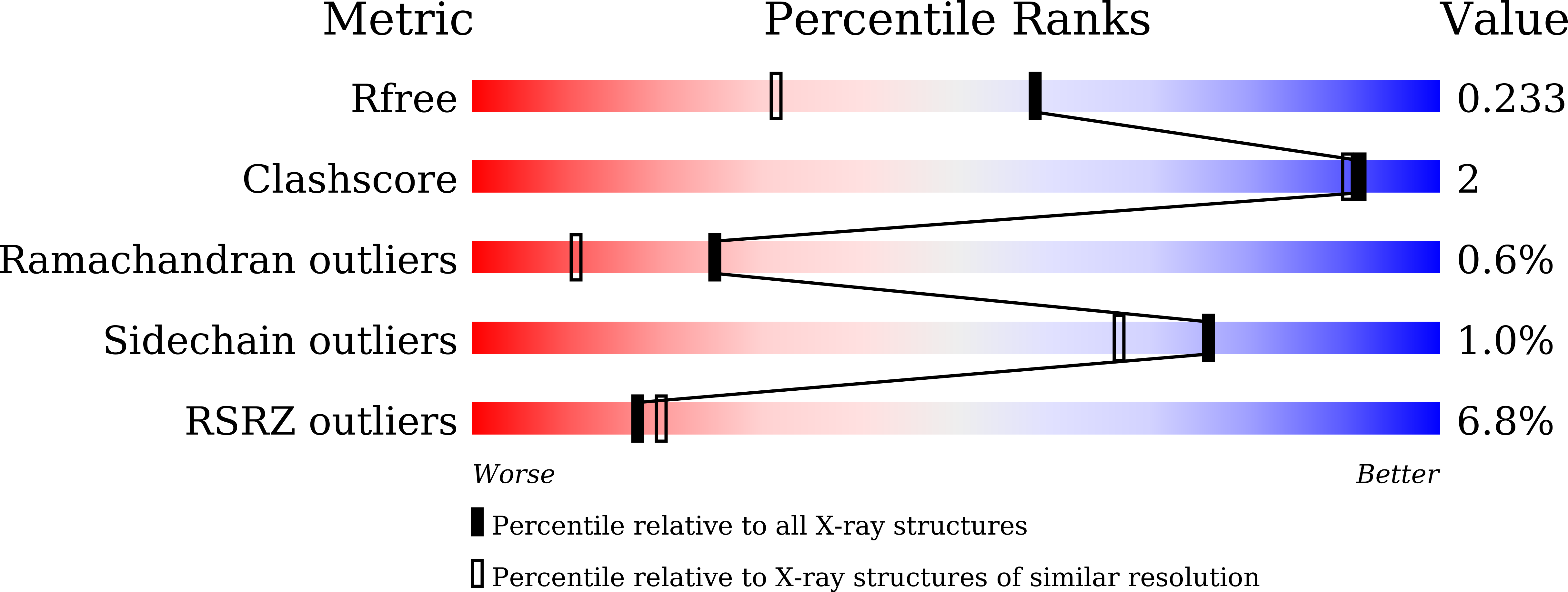
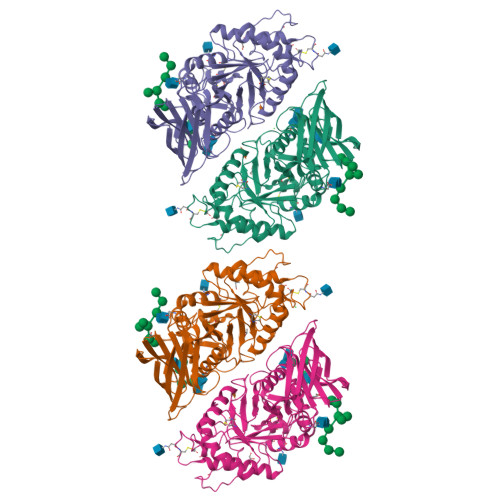
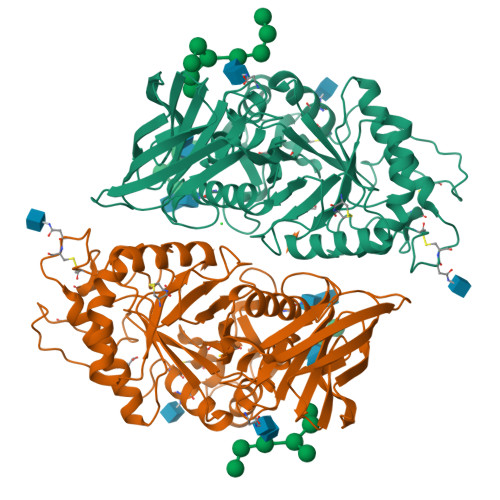
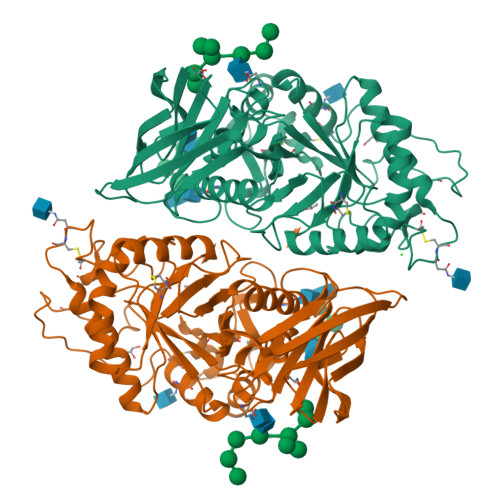
TcXyn30A from Talaromyces cellulolyticus, which belongs to subfamily 7 of the glycoside hydrolase family 30 (GH30-7), releases xylose from the reducing end of xylan and xylooligosaccharides (XOSs), the so-called reducing-end xylose-releasing exoxylanase (ReX). In this study, the crystal structures of TcXyn30A with and without xylose at subsite +1 (the binding site of the xylose residue at the reducing end) were determined. This is the first report on the structure of ReX in the family GH30-7. TcXyn30A forms a dimer. The complex structure of TcXyn30A with xylose revealed that subsite +1 is located at the dimer interface. TcXyn30A recognizes xylose at subsite +1 composed of amino acid residues from each monomer and blocks substrate binding to subsite +2 by dimer formation. Thus, the dimeric conformation is responsible for ReX activity. The structural comparison between TcXyn30A and the homologous enzyme indicated that subsite -2 is composed of assembled three stacked Trp residues, Trp49, Trp333, and Trp334, allowing TcXyn30A to accommodate xylan and any branched XOSs decorated with a substitution such as α-1,2-linked 4-O-methyl-d-glucuronic acid or α-1,2- and/or -1,3-linked L-arabinofuranose. These findings provide an insight into the structural determinants for ReX activity of TcXyn30A.
Organizational Affiliation:
Research Institute for Sustainable Chemistry, National Institute of Advanced Industrial Science and Technology (AIST), Higashi-Hiroshima, Hiroshima, Japan.