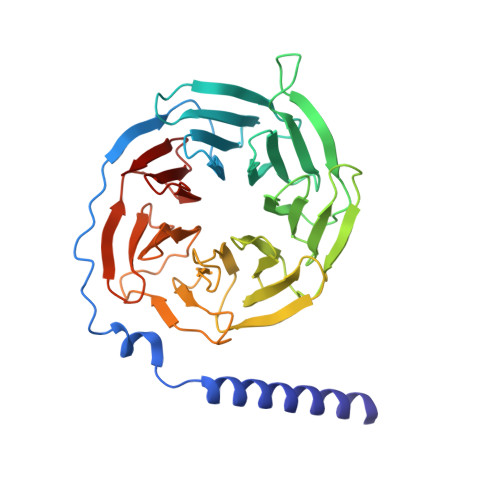
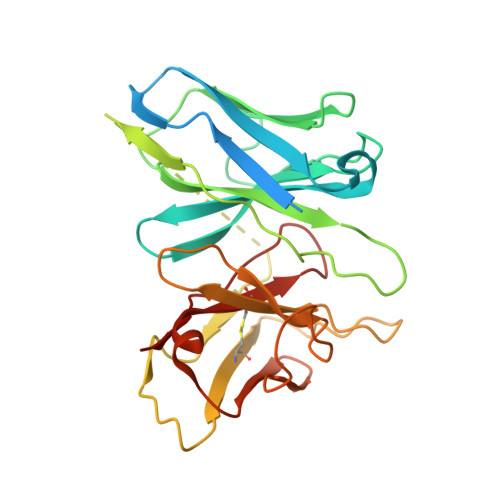
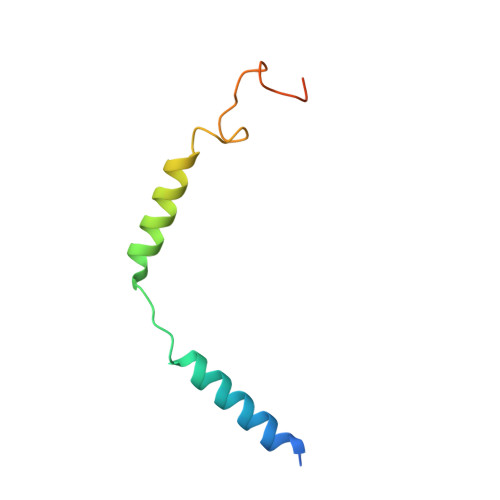
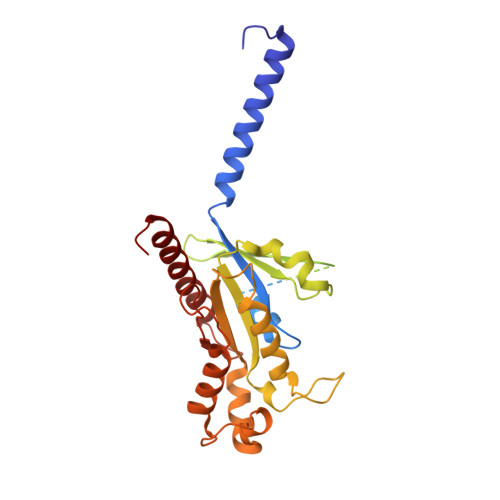
Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Mao, C., Xiao, P., Tao, X.N., Qin, J., He, Q.T., Zhang, C., Guo, S.C., Du, Y.Q., Chen, L.N., Shen, D.D., Yang, Z.S., Zhang, H.Q., Huang, S.M., He, Y.H., Cheng, J., Zhong, Y.N., Shang, P., Chen, J., Zhang, D.L., Wang, Q.L., Liu, M.X., Li, G.Y., Guo, Y., Xu, H.E., Wang, C., Zhang, C., Feng, S., Yu, X., Zhang, Y., Sun, J.P.(2023) Science 380: eadd6220-eadd6220
- PubMed: 36862765
- DOI: https://doi.org/10.1126/science.add6220
- Primary Citation of Related Structures:
8G59, 8ID3, 8ID4, 8ID6, 8ID8, 8ID9 - PubMed Abstract:
Individual free fatty acids (FAs) play important roles in metabolic homeostasis, many through engagement with more than 40G protein-coupled receptors. Searching for receptors to sense beneficial omega-3 FAs of fish oil enabled the identification of GPR120, which is involved in a spectrum of metabolic diseases. Here, we report six cryo-electron microscopy structures of GPR120 in complex with FA hormones or TUG891 and G i or G iq trimers. Aromatic residues inside the GPR120 ligand pocket were responsible for recognizing different double-bond positions of these FAs and connect ligand recognition to distinct effector coupling. We also investigated synthetic ligand selectivity and the structural basis of missense single-nucleotide polymorphisms. We reveal how GPR120 differentiates rigid double bonds and flexible single bonds. The knowledge gleaned here may facilitate rational drug design targeting to GPR120.
- Department of Biophysics and Department of Pathology of Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou 310058, China.
Organizational Affiliation: