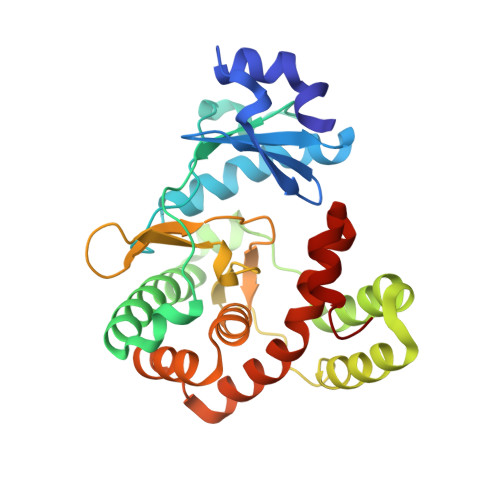
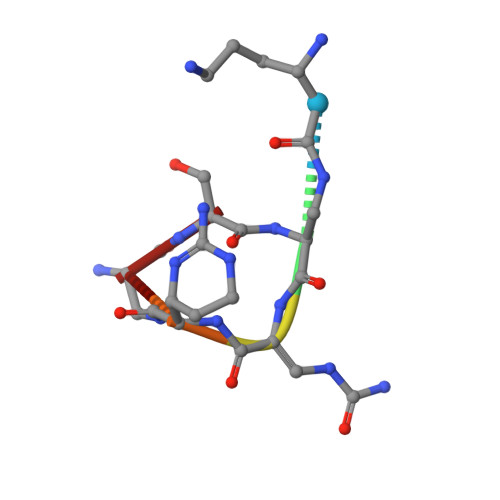
Discovery and characterization of genes conferring natural resistance to the antituberculosis antibiotic capreomycin.
Toh, S.I., Elaine Keisha, J., Wang, Y.L., Pan, Y.C., Jhu, Y.H., Hsiao, P.Y., Liao, W.T., Chen, P.Y., Ko, T.M., Chang, C.Y.(2023) Commun Biol 6: 1282-1282
- PubMed: 38114770
- DOI: https://doi.org/10.1038/s42003-023-05681-6
- Primary Citation of Related Structures:
8I80, 8I82, 8I84, 8I85, 8I86, 8I89, 8I8G, 8I8H - PubMed Abstract:
Metagenomic-based studies have predicted an extraordinary number of potential antibiotic-resistance genes (ARGs). These ARGs are hidden in various environmental bacteria and may become a latent crisis for antibiotic therapy via horizontal gene transfer. In this study, we focus on a resistance gene cph, which encodes a phosphotransferase (Cph) that confers resistance to the antituberculosis drug capreomycin (CMN). Sequence Similarity Network (SSN) analysis classified 353 Cph homologues into five major clusters, where the proteins in cluster I were found in a broad range of actinobacteria. We examine the function and antibiotics targeted by three putative resistance proteins in cluster I via biochemical and protein structural analysis. Our findings reveal that these three proteins in cluster I confer resistance to CMN, highlighting an important aspect of CMN resistance within this gene family. This study contributes towards understanding the sequence-structure-function relationships of the phosphorylation resistance genes that confer resistance to CMN.
Organizational Affiliation:
Department of Biological Science and Technology, National Yang Ming Chiao Tung University, Hsinchu, 30010, Taiwan, ROC.