Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Kamata, S., Honda, A., Ishikawa, R., Akahane, M., Fujita, A., Kaneko, C., Miyawaki, S., Habu, Y., Shiiyama, Y., Uchii, K., Machida, Y., Oyama, T., Ishii, I.(2023) Antioxidants (Basel) 12
- PubMed: 37627519
- DOI: https://doi.org/10.3390/antiox12081523
- Primary Citation of Related Structures:
8HUK, 8HUL, 8HUM, 8HUN, 8HUO, 8HUP, 8HUQ - PubMed Abstract:
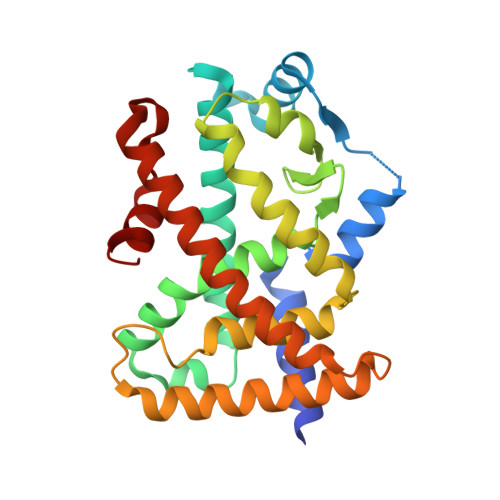
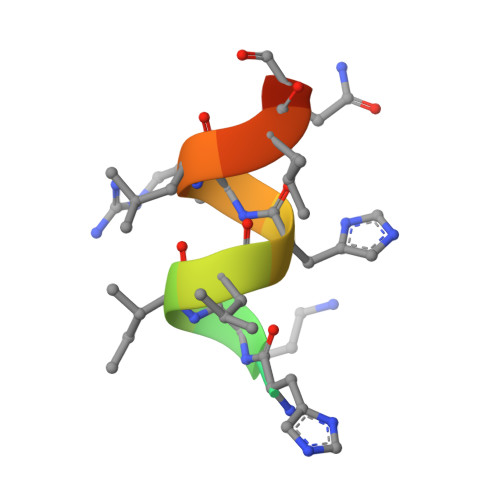
No therapeutic drugs are currently available for nonalcoholic steatohepatitis (NASH) that progresses from nonalcoholic fatty liver via oxidative stress-involved pathways. Three cognate peroxisome proliferator-activated receptor (PPAR) subtypes (PPARα/δ/γ) are considered as attractive targets. Although lanifibranor (PPARα/δ/γ pan agonist) and saroglitazar (PPARα/γ dual agonist) are currently under investigation in clinical trials for NASH, the development of seladelpar (PPARδ-selective agonist), elafibranor (PPARα/δ dual agonist), and many other dual/pan agonists has been discontinued due to serious side effects or little/no efficacies. This study aimed to obtain functional and structural insights into the potency, efficacy, and selectivity against PPARα/δ/γ of three current and past anti-NASH investigational drugs: lanifibranor, seladelpar, and elafibranor. Ligand activities were evaluated by three assays to detect different facets of the PPAR activation: transactivation assay, coactivator recruitment assay, and thermal stability assay. Seven high-resolution cocrystal structures (namely, those of the PPARα/δ/γ-ligand-binding domain (LBD)-lanifibranor, PPARα/δ/γ-LBD-seladelpar, and PPARα-LBD-elafibranor) were obtained through X-ray diffraction analyses, six of which represent the first deposit in the Protein Data Bank. Lanifibranor and seladelpar were found to bind to different regions of the PPARα/δ/γ-ligand-binding pockets and activated all PPAR subtypes with different potencies and efficacies in the three assays. In contrast, elafibranor induced transactivation and coactivator recruitment (not thermal stability) of all PPAR subtypes, but the PPARδ/γ-LBD-elafibranor cocrystals were not obtained. These results illustrate the highly variable PPARα/δ/γ activation profiles and binding modes of these PPAR ligands that define their pharmacological actions.
Organizational Affiliation:
Department of Health Chemistry, Showa Pharmaceutical University, Machida 194-8543, Tokyo, Japan.