Tighter ER retention of SARS-CoV-2 Omicron spike caused by a constellation of folding disruptive mutations
Li, Y.Q., Yang, M.R., Nan, Y.N., Wang, J.M., Guo, J.J., He, P.F., Dai, W.X., Zhou, S.Q., Wang, S.J., Zhang, Y., Ma, W.F.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protein transport protein Sec23A | 765 | Homo sapiens | Mutation(s): 0 Gene Names: SEC23A | 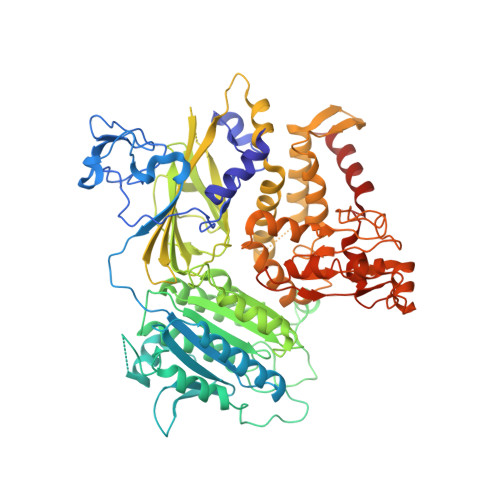 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q15436 (Homo sapiens) Explore Q15436 Go to UniProtKB: Q15436 | |||||
PHAROS: Q15436 GTEx: ENSG00000100934 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q15436 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protein transport protein Sec24A | 751 | Homo sapiens | Mutation(s): 0 Gene Names: SEC24A |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for O95486 (Homo sapiens) Explore O95486 Go to UniProtKB: O95486 | |||||
PHAROS: O95486 GTEx: ENSG00000113615 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O95486 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Vesicle-trafficking protein SEC22b | 199 | Homo sapiens | Mutation(s): 0 Gene Names: SEC22B, SEC22L1 |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for O75396 (Homo sapiens) Explore O75396 Go to UniProtKB: O75396 | |||||
PHAROS: O75396 GTEx: ENSG00000265808 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O75396 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| HCoV-OC43 | 10 | Human coronavirus OC43 | Mutation(s): 0 |  | |
UniProt | |||||
Find proteins for P36334 (Human coronavirus OC43) Explore P36334 Go to UniProtKB: P36334 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P36334 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ZN Query on ZN | E [auth A], F [auth B] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 148.58 | α = 90 |
| b = 96.203 | β = 90.01 |
| c = 130.465 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-3000 | data reduction |
| SCALA | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Beijing University of Chinese Medicine (BUCM) | China | 90011451310011, 1000061223476, ZYYCXTD-C-202006 |