Structural basis of human Slo2.2 channel gating and modulation.
Zhang, J., Liu, S., Fan, J., Yan, R., Huang, B., Zhou, F., Yuan, T., Gong, J., Huang, Z., Jiang, D.(2023) Cell Rep 42: 112858-112858
- PubMed: 37494189
- DOI: https://doi.org/10.1016/j.celrep.2023.112858
- Primary Citation of Related Structures:
8HIR, 8HK6, 8HKF, 8HKK, 8HKM, 8HKQ - PubMed Abstract:
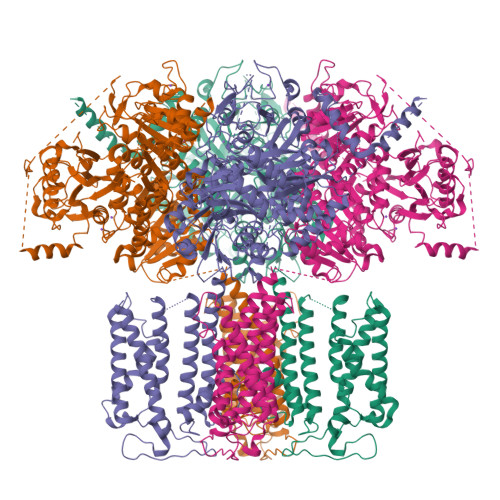
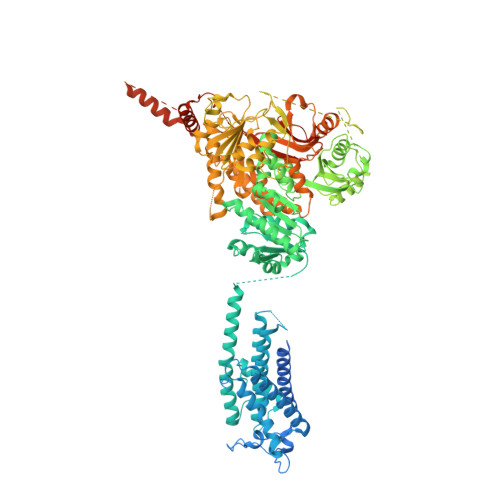
The sodium-activated Slo2.2 channel is abundantly expressed in the brain, playing a critical role in regulating neuronal excitability. The Na + -binding site and the underlying mechanisms of Na + -dependent activation remain unclear. Here, we present cryoelectron microscopy (cryo-EM) structures of human Slo2.2 in closed, open, and inhibitor-bound form at resolutions of 2.6-3.2 Å, revealing gating mechanisms of Slo2.2 regulation by cations and a potent inhibitor. The cytoplasmic gating ring domain of the closed Slo2.2 harbors multiple K + and Zn 2+ sites, which stabilize the channel in the closed conformation. The open Slo2.2 structure reveals at least two Na + -sensitive sites where Na + binding induces expansion and rotation of the gating ring that opens the inner gate. Furthermore, a potent inhibitor wedges into a pocket formed by pore helix and S6 helix and blocks the pore. Together, our results provide a comprehensive structural framework for the investigation of Slo2.2 channel gating, Na + sensation, and inhibition.
Organizational Affiliation:
Laboratory of Soft Matter Physics, Institute of Physics, Chinese Academy of Sciences, Beijing 100190, China; College of Life Science and Technology, Key Laboratory of Molecular Biophysics of MOE, Huazhong University of Science and Technology, Wuhan, Hubei, China.