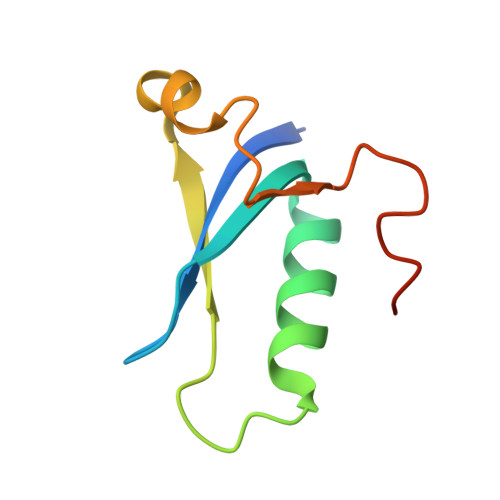
The structure of AcrIC9 revealing the putative inhibitory mechanism of AcrIC9 against the type IC CRISPR-Cas system.
Kang, Y.J., Kim, J.H., Lee, G.H., Ha, H.J., Park, Y.H., Hong, E., Park, H.H.(2023) IUCrJ 10: 624-634
- PubMed: 37668219
- DOI: https://doi.org/10.1107/S2052252523007236
- Primary Citation of Related Structures:
8HJJ - PubMed Abstract:
CRISPR-Cas systems are known to be part of the bacterial adaptive immune system that provides resistance against intruders such as viruses, phages and other mobile genetic elements. To combat this bacterial defense mechanism, phages encode inhibitors called Acrs (anti-CRISPR proteins) that can suppress them. AcrIC9 is the most recently identified member of the AcrIC family that inhibits the type IC CRISPR-Cas system. Here, the crystal structure of AcrIC9 from Rhodobacter capsulatus is reported, which comprises a novel fold made of three central antiparallel β-strands surrounded by three α-helixes, a structure that has not been detected before. It is also shown that AcrIC9 can form a dimer via disulfide bonds generated by the Cys69 residue. Finally, it is revealed that AcrIC9 directly binds to the type IC cascade. Analysis and comparison of its structure with structural homologs indicate that AcrIC9 belongs to DNA-mimic Acrs that directly bind to the cascade complex and hinder the target DNA from binding to the cascade.
Organizational Affiliation:
College of Pharmacy, Chung-Ang University, Seoul 06974, Republic of Korea.