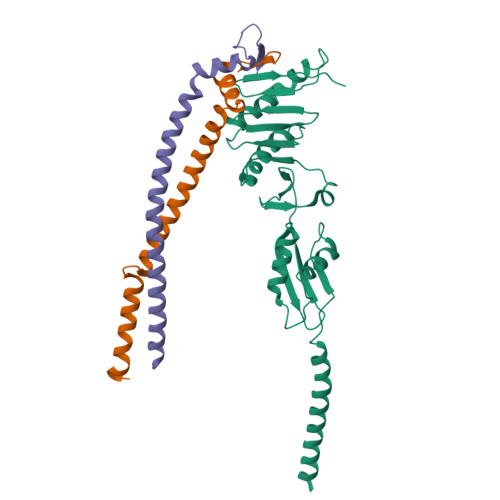
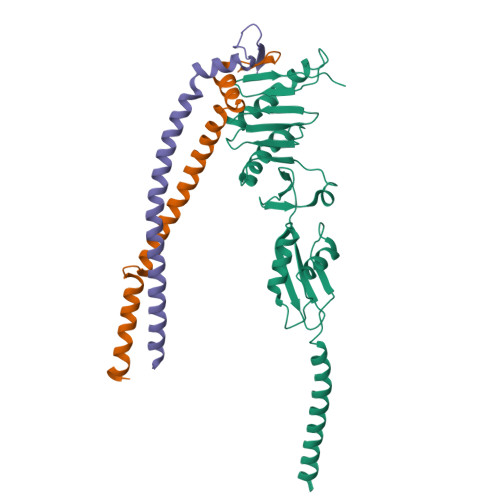
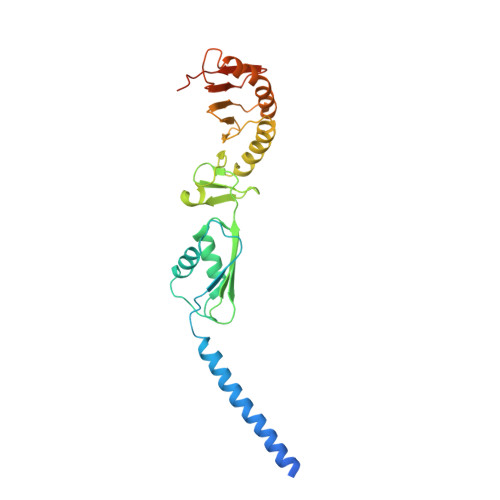
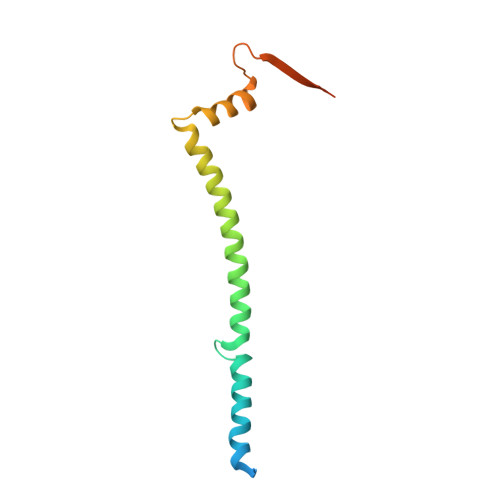
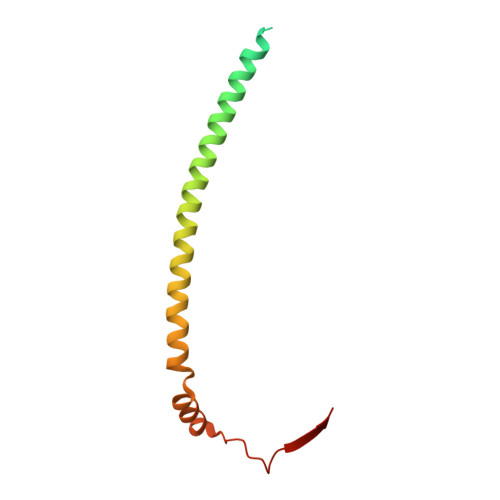
Structure of the heterotrimeric membrane protein complex FtsB-FtsL-FtsQ of the bacterial divisome.
Nguyen, H.T.V., Chen, X., Parada, C., Luo, A.C., Shih, O., Jeng, U.S., Huang, C.Y., Shih, Y.L., Ma, C.(2023) Nat Commun 14: 1903-1903
- PubMed: 37019934
- DOI: https://doi.org/10.1038/s41467-023-37543-4
- Primary Citation of Related Structures:
8HHF, 8HHG, 8HHH - PubMed Abstract:
The synthesis of the cell-wall peptidoglycan during bacterial cell division is mediated by a multiprotein machine, called the divisome. The essential membrane protein complex of FtsB, FtsL and FtsQ (FtsBLQ) is at the heart of the divisome assembly cascade in Escherichia coli. This complex regulates the transglycosylation and transpeptidation activities of the FtsW-FtsI complex and PBP1b via coordination with FtsN, the trigger for the onset of constriction. Yet the underlying mechanism of FtsBLQ-mediated regulation is largely unknown. Here, we report the full-length structure of the heterotrimeric FtsBLQ complex, which reveals a V-shaped architecture in a tilted orientation. Such a conformation could be strengthened by the transmembrane and the coiled-coil domains of the FtsBL heterodimer, as well as an extended β-sheet of the C-terminal interaction site involving all three proteins. This trimeric structure may also facilitate interactions with other divisome proteins in an allosteric manner. These results lead us to propose a structure-based model that delineates the mechanism of the regulation of peptidoglycan synthases by the FtsBLQ complex.
Organizational Affiliation:
Genomics Research Center, Academia Sinica, Taipei, 115, Taiwan.