Foot-and-mouth disease virus antigenic landscape and reduced immunogenicity elucidated in atomic detail.
Li, H., Liu, P., Dong, H., Dekker, A., Harmsen, M.M., Guo, H., Wang, X., Sun, S.(2024) Nat Commun 15: 8774-8774
- PubMed: 39389971
- DOI: https://doi.org/10.1038/s41467-024-53027-5
- Primary Citation of Related Structures:
8HBG, 8HBI, 8HBJ, 8HEE, 8HEG - PubMed Abstract:
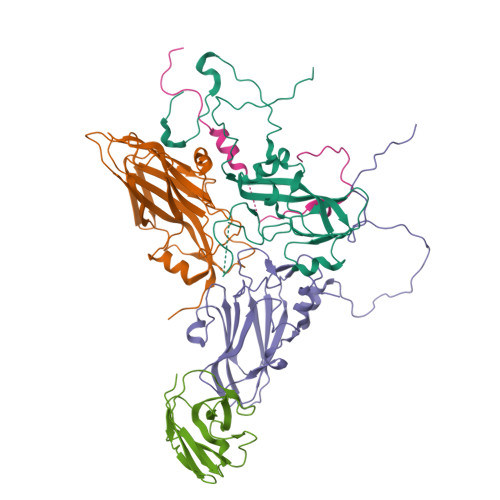
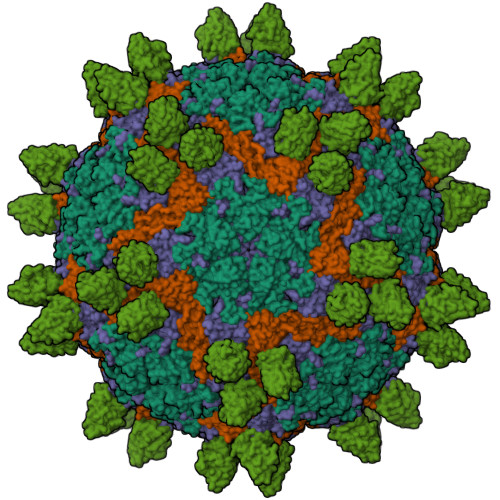
Unlike most other picornaviruses, foot-and-mouth disease (FMD) intact virions (146S) dissociate easily into small pentameric subunits (12S). This causes a dramatically decreased immunogenicity by a mechanism that remains elusive. Here, we present the high-resolution structures of 12S (3.2 Å) and its immune complex of a single-domain antibody (VHH) targeting the particle interior (3.2 Å), as well as two 146S-specific VHHs complexed to distinct sites on the 146S capsid surface (3.6 Å and 2.9 Å). The antigenic landscape of 146S is depicted using 13 known FMD virus-antibody complexes. Comparison of the immunogenicity of 146S and 12S in pigs, focusing on the resulting antigenic sites and incorporating structural analysis, reveals that dissociation of 146S leads to structural alteration and destruction of multiple epitopes, resulting in significant differences in antibody profiles/lineages induced by 12S and 146S. Furthermore, 146S generates higher synergistic neutralizing antibody titers compared to 12S, whereas both particles induce similar total FMD virus specific antibody titers. This study can guide the structure-based rational design of novel multivalent and broad-spectrum recombinant vaccines for protection against FMD.
Organizational Affiliation:
State Key Laboratory for Animal Disease Control and Prevention, College of Veterinary Medicine, Lanzhou University, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Lanzhou, China.