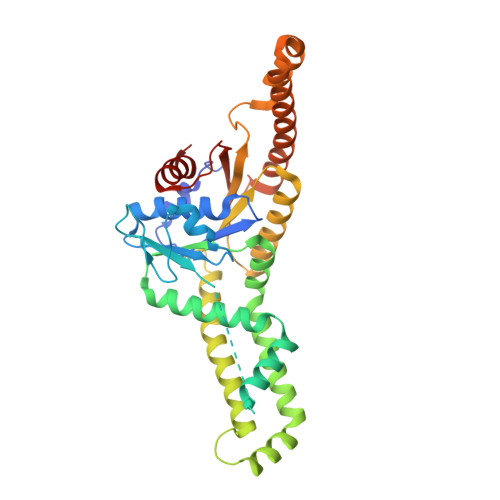
A distinct dimer configuration of a diatom Get3 forming a tetrameric complex with its tail-anchored membrane cargo.
Chen, C.C., Huang, Y.R., Chan, Y.T., Lin, H.Y., Lin, H.J., Hsiao, C.D., Ko, T.P., Lin, T.W., Lan, Y.H., Lin, H.Y., Chang, H.Y.(2024) BMC Biol 22: 136-136
- PubMed: 38867239
- DOI: https://doi.org/10.1186/s12915-024-01933-x
- Primary Citation of Related Structures:
8HAC, 8HAD - PubMed Abstract:
Most tail-anchored (TA) membrane proteins are delivered to the endoplasmic reticulum through a conserved posttranslational pathway. Although core mechanisms underlying the targeting and insertion of TA proteins are well established in eukaryotes, their role in mediating TA protein biogenesis in plants remains unclear. We reported the crystal structures of algal arsenite transporter 1 (ArsA1), which possesses an approximately 80-kDa monomeric architecture and carries chloroplast-localized TA proteins. However, the mechanistic basis of ArsA2, a Get3 (guided entry of TA proteins 3) homolog in plants, for TA recognition remains unknown. Here, for the first time, we present the crystal structures of the diatom Pt-Get3a that forms a distinct ellipsoid-shaped tetramer in the open (nucleotide-bound) state through crystal packing. Pulldown assay results revealed that only tetrameric Pt-Get3a can bind to TA proteins. The lack of the conserved zinc-coordination CXXC motif in Pt-Get3a potentially leads to the spontaneous formation of a distinct parallelogram-shaped dimeric conformation in solution, suggesting a new dimer state for subsequent tetramerization upon TA targeting. Pt-Get3a nonspecifically binds to different subsets of TA substrates due to the lower hydrophobicity of its α-helical subdomain, which is implicated in TA recognition. Our study provides new insights into the mechanisms underlying TA protein shielding by tetrameric Get3 during targeting to the diatom's cell membrane.
Organizational Affiliation:
Department of Life Sciences and Institute of Genome Sciences, National Yang Ming Chiao Tung University, Beitou Dist, No. 155, Sec. 2, Linong St, Taipei City, 112304, Taiwan.