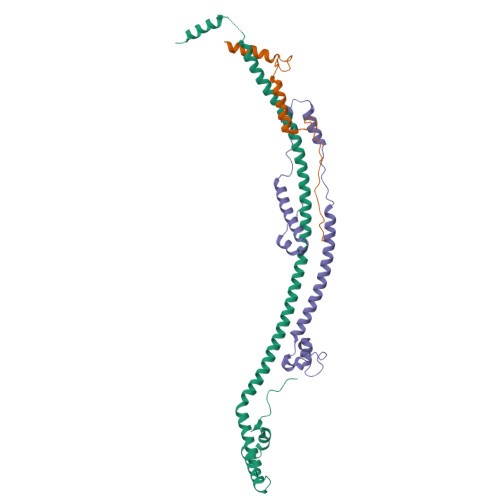
Structure of the human ATP synthase.
Lai, Y., Zhang, Y., Zhou, S., Xu, J., Du, Z., Feng, Z., Yu, L., Zhao, Z., Wang, W., Tang, Y., Yang, X., Guddat, L.W., Liu, F., Gao, Y., Rao, Z., Gong, H.(2023) Mol Cell 83: 2137
- PubMed: 37244256
- DOI: https://doi.org/10.1016/j.molcel.2023.04.029
- Primary Citation of Related Structures:
8H9E, 8H9F, 8H9G, 8H9I, 8H9J, 8H9K, 8H9L, 8H9M, 8H9N, 8H9P, 8H9Q, 8H9R, 8H9S, 8H9T, 8H9U, 8H9V - PubMed Abstract:
Biological energy currency ATP is produced by F 1 F o -ATP synthase. However, the molecular mechanism for human ATP synthase action remains unknown. Here, we present snapshot images for three main rotational states and one substate of human ATP synthase using cryoelectron microscopy. These structures reveal that the release of ADP occurs when the β subunit of F 1 F o -ATP synthase is in the open conformation, showing how ADP binding is coordinated during synthesis. The accommodation of the symmetry mismatch between F 1 and F o motors is resolved by the torsional flexing of the entire complex, especially the γ subunit, and the rotational substep of the c subunit. Water molecules are identified in the inlet and outlet half-channels, suggesting that the proton transfer in these two half-channels proceed via a Grotthus mechanism. Clinically relevant mutations are mapped to the structure, showing that they are mainly located at the subunit-subunit interfaces, thus causing instability of the complex.
Organizational Affiliation:
State Key Laboratory of Medicinal Chemical Biology and Frontiers Science Center for Cell Responses, College of Life Sciences, Nankai University, Tianjin 300071, China; Institute for Immunology, College of Life Sciences, Nankai University, Tianjin 300071, China.