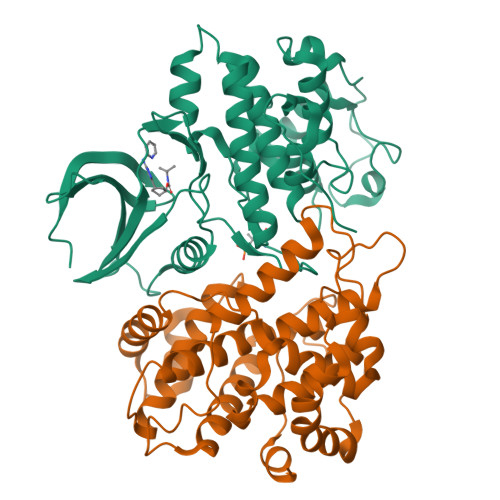
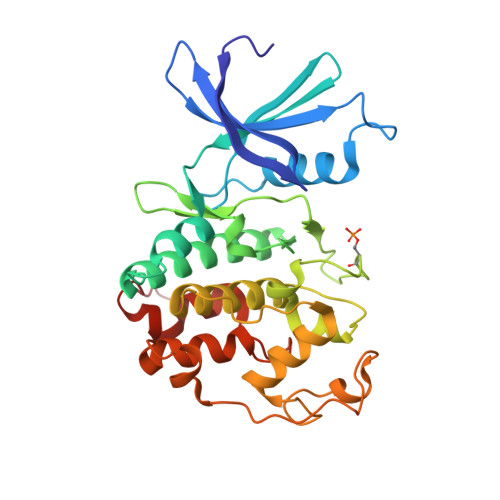
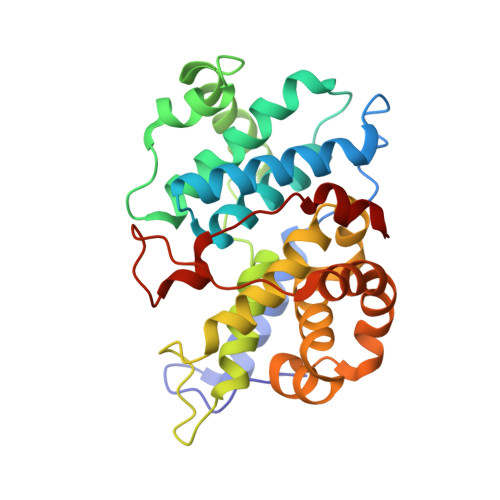
Accelerated Discovery of Macrocyclic CDK2 Inhibitor QR-6401 by Generative Models and Structure-Based Drug Design.
Yu, Y., Huang, J., He, H., Han, J., Ye, G., Xu, T., Sun, X., Chen, X., Ren, X., Li, C., Li, H., Huang, W., Liu, Y., Wang, X., Gao, Y., Cheng, N., Guo, N., Chen, X., Feng, J., Hua, Y., Liu, C., Zhu, G., Xie, Z., Yao, L., Zhong, W., Chen, X., Liu, W., Li, H.(2023) ACS Med Chem Lett 14: 297-304
- PubMed: 36923916
- DOI: https://doi.org/10.1021/acsmedchemlett.2c00515
- Primary Citation of Related Structures:
8H6P, 8H6T - PubMed Abstract:
Selective CDK2 inhibitors have the potential to provide effective therapeutics for CDK2-dependent cancers and for combating drug resistance due to high cyclin E1 (CCNE1) expression intrinsically or CCNE1 amplification induced by treatment of CDK4/6 inhibitors. Generative models that take advantage of deep learning are being increasingly integrated into early drug discovery for hit identification and lead optimization. Here we report the discovery of a highly potent and selective macrocyclic CDK2 inhibitor QR-6401 ( 23 ) accelerated by the application of generative models and structure-based drug design (SBDD). QR-6401 ( 23 ) demonstrated robust antitumor efficacy in an OVCAR3 ovarian cancer xenograft model via oral administration.
Organizational Affiliation:
Tencent AI Lab, Tencent, Shenzhen 518057, China.