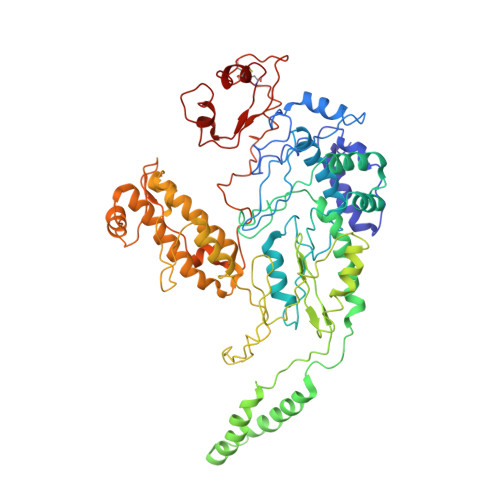
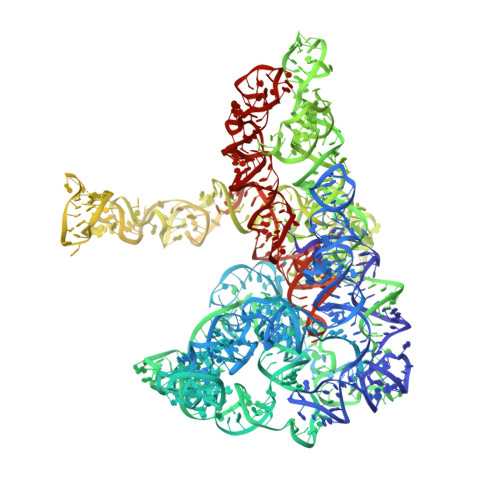
Functionalized graphene grids with various charges for single-particle cryo-EM.
Lu, Y., Liu, N., Liu, Y., Zheng, L., Yang, J., Wang, J., Jia, X., Zi, Q., Peng, H., Rao, Y., Wang, H.W.(2022) Nat Commun 13: 6718-6718
- PubMed: 36344519
- DOI: https://doi.org/10.1038/s41467-022-34579-w
- Primary Citation of Related Structures:
8H2H - PubMed Abstract:
A major hurdle for single particle cryo-EM in structural determination lies in the specimen preparation impaired by the air-water interface (AWI) and preferential particle-orientation problems. In this work, we develop functionalized graphene grids with various charges via a dediazoniation reaction for cryo-EM specimen preparation. The graphene grids are paraffin-assistant fabricated, which appear with less contaminations compared with those produced by polymer transfer method. By applying onto three different types of macromolecules, we demonstrate that the high-yield charged graphene grids bring macromolecules away from the AWI and enable adjustable particle-orientation distribution for more robust single particle cryo-EM structural determination.
Organizational Affiliation:
School of Life Sciences, Tsinghua University, Beijing, China.