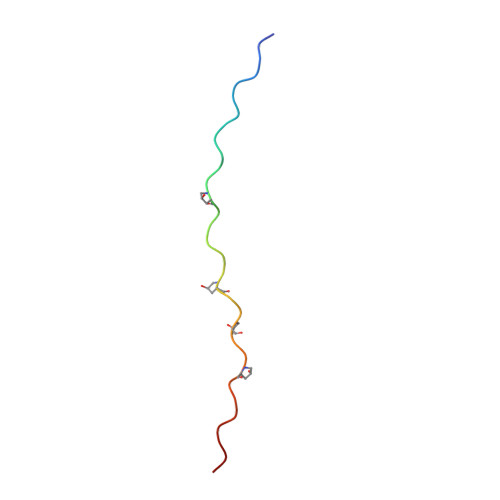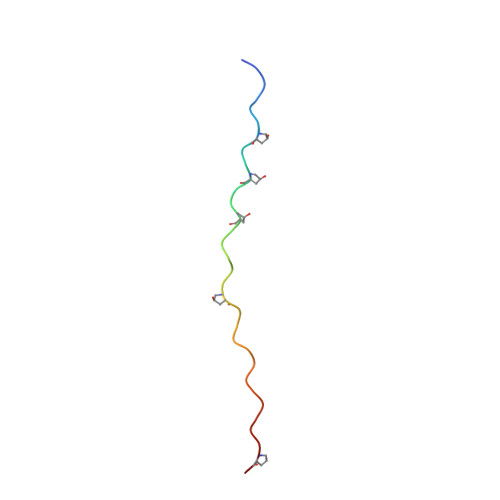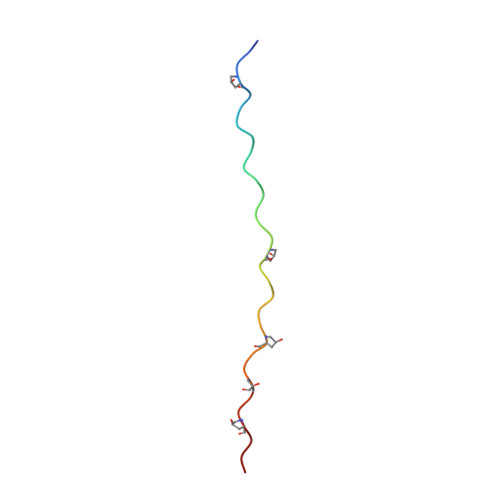Stability of collagen heterotrimer with same charge pattern and different charged residue identities.
Huang, Y., Lan, J., Wu, C., Zhang, R., Zheng, H., Fan, S., Xu, F.(2023) Biophys J 122: 2686-2695
- PubMed: 37226442
- DOI: https://doi.org/10.1016/j.bpj.2023.05.023
- Primary Citation of Related Structures:
8GZO, 8H0E, 8H0F - PubMed Abstract:
Salt bridges are important factors in maintaining the stability of proteins, and their contribution to protein folding has received much attention. Although the interaction energies, or stabilizing contributions, of individual salt bridges have been measured in various proteins, a systematic assessment of various types of salt bridges in a relatively uniform environment is still a valuable analysis. Here, we used a collagen heterotrimer as a host-guest platform to construct 48 heterotrimers with the same charge pattern. A variety of salt bridges were formed between the oppositely charged residues Lys, Arg, Asp, and Glu. The melting temperature (T m ) of the heterotrimers was measured with circular dichroism. The atomic structures of 10 salt bridges were shown in three x-ray crystals of heterotrimer. Molecular dynamics simulation based on the crystal structures indicated that strong, intermediate, and weak salt bridges have distinctive N-O distances. A linear regression model was used to predict the stability of heterotrimers with high accuracy (R 2 = 0.93). We developed an online database to help readers understand how a salt bridge stabilizes collagen. This work will help us better understand the stabilizing mechanism of salt bridges in collagen folding and provide a new strategy to design collagen heterotrimers.
Organizational Affiliation:
Ministry of Education Key Laboratory of Industrial Biotechnology, School of Biotechnology, Jiangnan University, Wuxi, China.