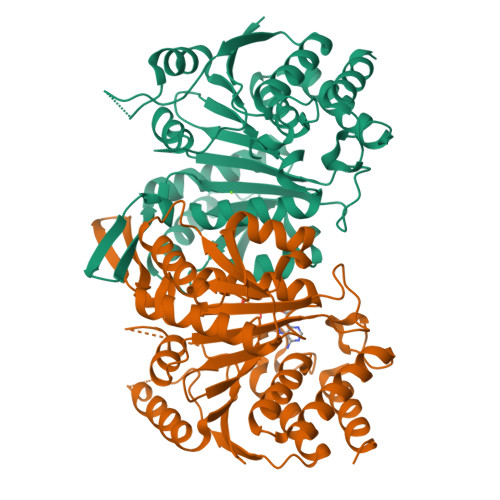
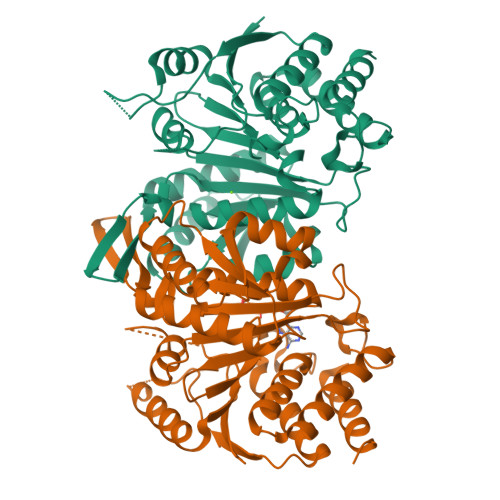
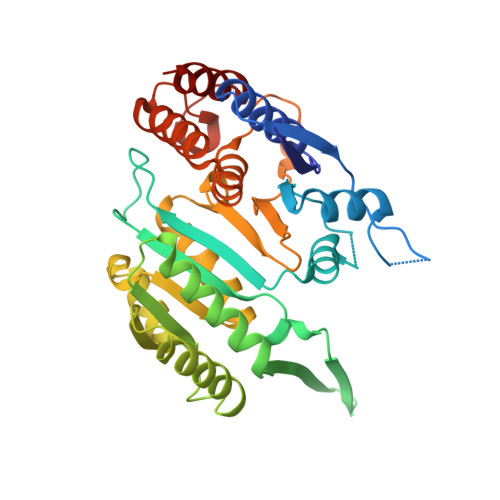
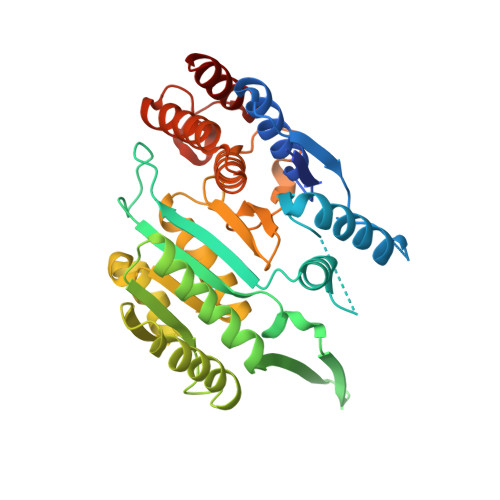
Structures of a constitutively active mutant of human IDH3 reveal new insights into the mechanisms of allosteric activation and the catalytic reaction.
Chen, X., Sun, P., Liu, Y., Shen, S., Ma, T., Ding, J.(2022) J Biological Chem 298: 102695-102695
- PubMed: 36375638
- DOI: https://doi.org/10.1016/j.jbc.2022.102695
- Primary Citation of Related Structures:
8GRB, 8GRD, 8GRG, 8GRH, 8GRU, 8GS5 - PubMed Abstract:
Human NAD-dependent isocitrate dehydrogenase or IDH3 (HsIDH3) catalyzes the decarboxylation of isocitrate into α-ketoglutarate in the tricarboxylic acid cycle. It consists of three types of subunits (α, β, and γ) and exists and functions as the (αβαγ) 2 heterooctamer. HsIDH3 is regulated allosterically and/or competitively by numerous metabolites including CIT, ADP, ATP, and NADH. Our previous studies have revealed the molecular basis for the activity and regulation of the αβ and αγ heterodimers. However, the molecular mechanism for the allosteric activation of the HsIDH3 holoenzyme remains elusive. In this work, we report the crystal structures of the αβ and αγ heterodimers and the (αβαγ) 2 heterooctamer containing an α-Q139A mutation in the clasp domain, which renders all the heterodimers and the heterooctamer constitutively active in the absence of activators. Our structural analysis shows that the α-Q139A mutation alters the hydrogen-bonding network at the heterodimer-heterodimer interface in a manner similar to that in the activator-bound αγ heterodimer. This alteration not only stabilizes the active sites of both α Q139A β and α Q139A γ heterodimers in active conformations but also induces conformational changes of the pseudo-allosteric site of the α Q139A β heterodimer enabling it to bind activators. In addition, the α Q139A ICT+Ca+NAD β NAD structure presents the first pseudo-Michaelis complex of HsIDH3, which allows us to identify the key residues involved in the binding of cofactor, substrate, and metal ion. Our structural and biochemical data together reveal new insights into the molecular mechanisms for allosteric regulation and the catalytic reaction of HsIDH3.
Organizational Affiliation:
State Key Laboratory of Molecular Biology, Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, University of Chinese Academy of Sciences, Chinese Academy of Sciences, Shanghai, China.