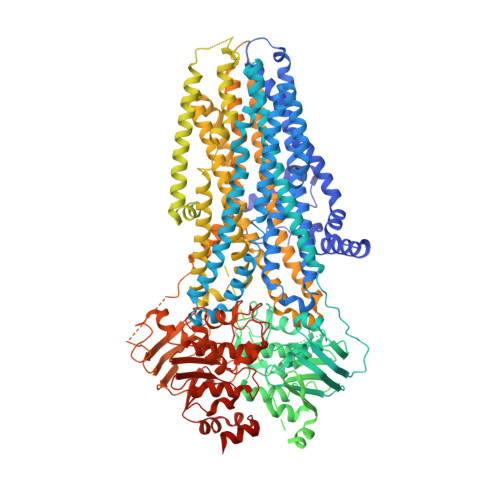
Structure-based discovery of positive and negative CFTR modulators
Liu, F., Kaplan, A.L., Levring, J., Einsiedel, J., Tiedt, S., Distler, K., Omattage, N.S., Kondratov, I.S., Moroz, Y.S., Irwin, J.J., Gmeiner, P., Shoichet, B.K., Chen, J.To be published.