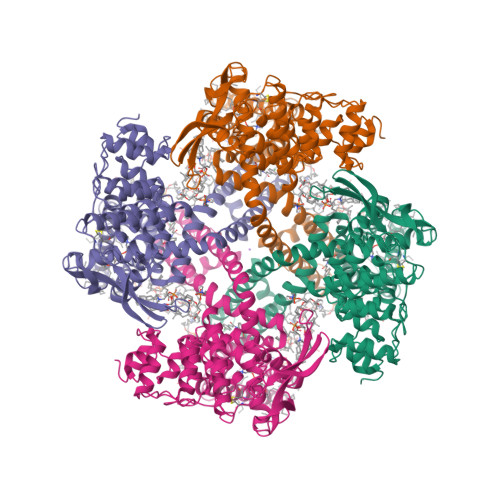
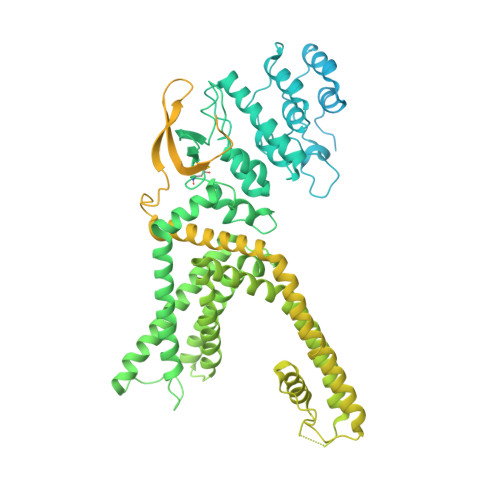
Human TRPV1 structure and inhibition by the analgesic SB-366791.
Neuberger, A., Oda, M., Nikolaev, Y.A., Nadezhdin, K.D., Gracheva, E.O., Bagriantsev, S.N., Sobolevsky, A.I.(2023) Nat Commun 14: 2451-2451
- PubMed: 37117175
- DOI: https://doi.org/10.1038/s41467-023-38162-9
- Primary Citation of Related Structures:
8GF8, 8GF9, 8GFA - PubMed Abstract:
Pain therapy has remained conceptually stagnant since the opioid crisis, which highlighted the dangers of treating pain with opioids. An alternative addiction-free strategy to conventional painkiller-based treatment is targeting receptors at the origin of the pain pathway, such as transient receptor potential (TRP) ion channels. Thus, a founding member of the vanilloid subfamily of TRP channels, TRPV1, represents one of the most sought-after pain therapy targets. The need for selective TRPV1 inhibitors extends beyond pain treatment, to other diseases associated with this channel, including psychiatric disorders. Here we report the cryo-electron microscopy structures of human TRPV1 in the apo state and in complex with the TRPV1-specific nanomolar-affinity analgesic antagonist SB-366791. SB-366791 binds to the vanilloid site and acts as an allosteric hTRPV1 inhibitor. SB-366791 binding site is supported by mutagenesis combined with electrophysiological recordings and can be further explored to design new drugs targeting TRPV1 in disease conditions.
Organizational Affiliation:
Department of Biochemistry and Molecular Biophysics, Columbia University, New York, NY, USA.