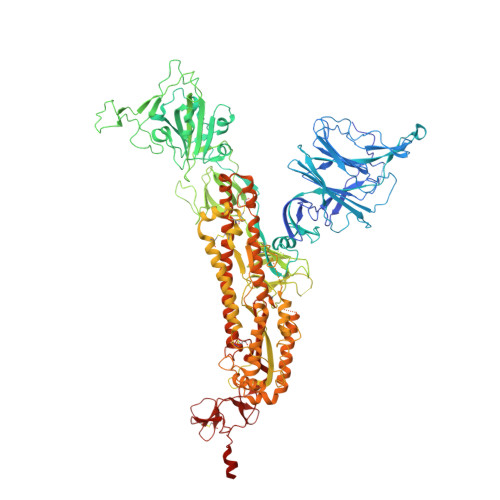
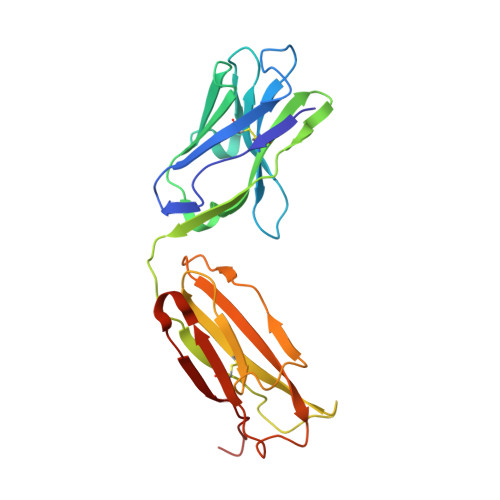
Elucidating the mechanism of SARS-CoV-2 Omicron variant escape from a RBD class-3 human antibody
Patel, A., Kumar, S., Lai, L., Chakravarthy, C., Valanparambil, R., Keen, M., Laughlin, Z.T., Frank, F., Cheedarla, N., Verkerke, H.P., Neish, A.S., Roback, J.D., Davis, C.W., Wrammert, J., Ahmed, R., Suthar, M.S., Murali-Krishna, K., Chandele, A., Ortlund, E.A.To be published.