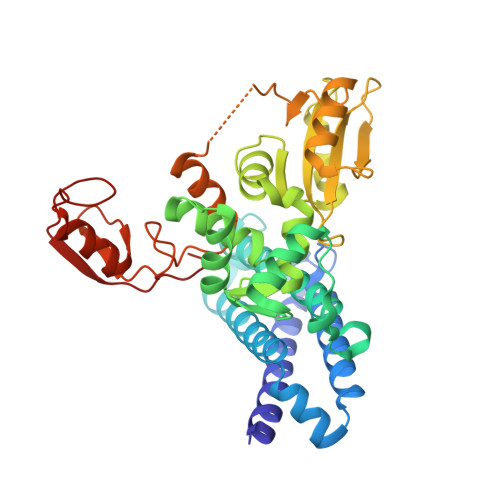
The co-crystal structure of Cbl-b and a small-molecule inhibitor reveals the mechanism of Cbl-b inhibition.
Kimani, S.W., Perveen, S., Szewezyk, M., Zeng, H., Dong, A., Li, F., Ghiabi, P., Li, Y., Chau, I., Arrowsmith, C.H., Barsyte-Lovejoy, D., Santhakumar, V., Vedadi, M., Halabelian, L.(2023) Commun Biol 6: 1272-1272
- PubMed: 38104184
- DOI: https://doi.org/10.1038/s42003-023-05655-8
- Primary Citation of Related Structures:
8GCY - PubMed Abstract:
Cbl-b is a RING-type E3 ubiquitin ligase that is expressed in several immune cell lineages, where it negatively regulates the activity of immune cells. Cbl-b has specifically been identified as an attractive target for cancer immunotherapy due to its role in promoting an immunosuppressive tumor environment. A Cbl-b inhibitor, Nx-1607, is currently in phase I clinical trials for advanced solid tumor malignancies. Using a suite of biophysical and cellular assays, we confirm potent binding of C7683 (an analogue of Nx-1607) to the full-length Cbl-b and its N-terminal fragment containing the TKBD-LHR-RING domains. To further elucidate its mechanism of inhibition, we determined the co-crystal structure of Cbl-b with C7683, revealing the compound's interaction with both the TKBD and LHR, but not the RING domain. Here, we provide structural insights into a novel mechanism of Cbl-b inhibition by a small-molecule inhibitor that locks the protein in an inactive conformation by acting as an intramolecular glue.
Organizational Affiliation:
Structural Genomics Consortium, University of Toronto, Toronto, ON, Canada.