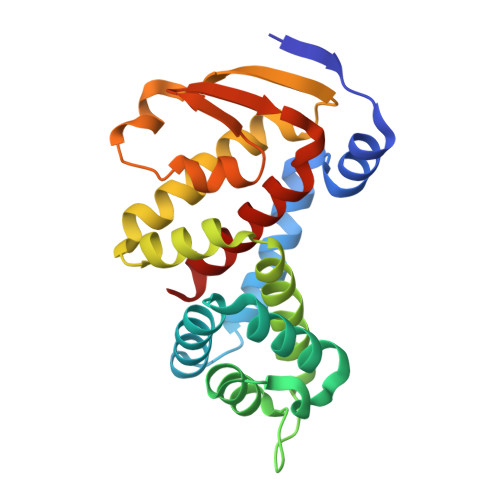
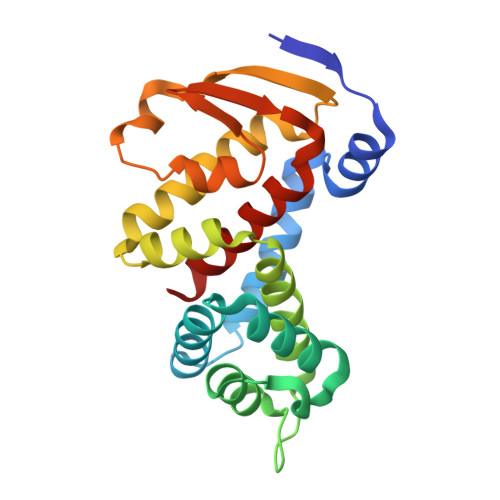
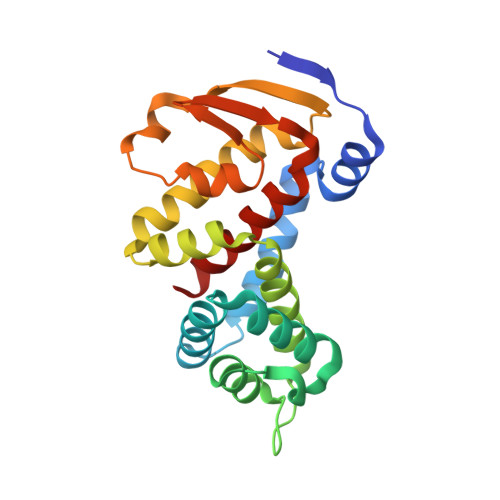
Structural homology screens reveal poxvirus-encoded proteins impacting inflammasome-mediated defenses.
Boys, I.N., Johnson, A.G., Quinlan, M., Kranzusch, P.J., Elde, N.C.(2023) bioRxiv
- PubMed: 36909515
- DOI: https://doi.org/10.1101/2023.02.26.529821
- Primary Citation of Related Structures:
8GBE - PubMed Abstract:
Viruses acquire host genes via horizontal gene transfer and can express them to manipulate host biology during infections. Some viral and host homologs retain sequence identity, but evolutionary divergence can obscure host origins. We used structural modeling to compare vaccinia virus proteins with metazoan proteomes. We identified vaccinia A47L as a homolog of gasdermins, the executioners of pyroptosis. An X-ray crystal structure of A47 confirmed this homology and cell-based assays revealed that A47 inhibits pyroptosis. We also identified vaccinia C1L as the product of a cryptic gene fusion event coupling a Bcl-2 related fold with a pyrin domain. C1 associates with components of the inflammasome, a cytosolic innate immune sensor involved in pyroptosis, yet paradoxically enhances inflammasome activity, suggesting a benefit to poxvirus replication in some circumstances. Our findings demonstrate the potential of structural homology screens to reveal genes that viruses capture from hosts and repurpose to benefit viral fitness.
Organizational Affiliation:
Department of Human Genetics, University of Utah, Salt Lake City, Utah, 84112 USA.