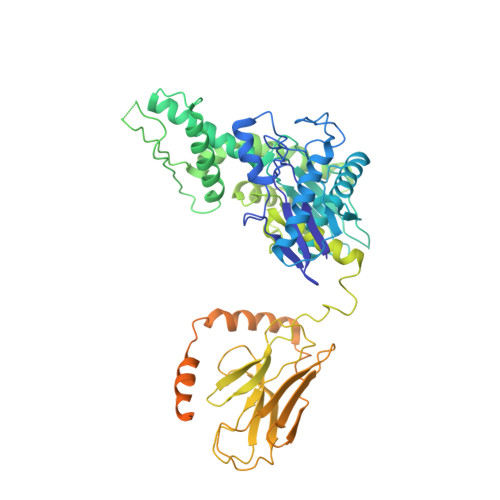
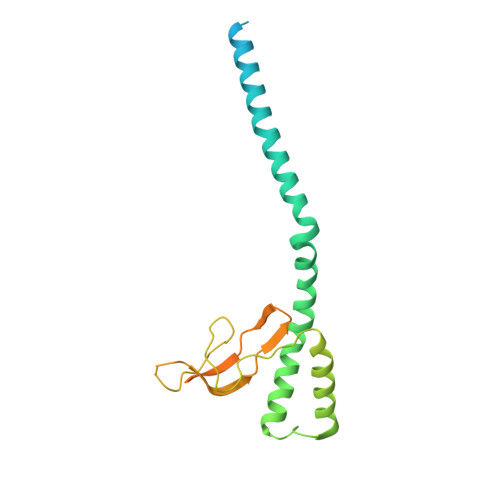
Structure of the M. tuberculosis DnaK-GrpE complex reveals how key DnaK roles are controlled.
Xiao, X., Fay, A., Molina, P.S., Kovach, A., Glickman, M.S., Li, H.(2024) Nat Commun 15: 660-660
- PubMed: 38253530
- DOI: https://doi.org/10.1038/s41467-024-44933-9
- Primary Citation of Related Structures:
8GB3 - PubMed Abstract:
The molecular chaperone DnaK is essential for viability of Mycobacterium tuberculosis (Mtb). DnaK hydrolyzes ATP to fold substrates, and the resulting ADP is exchanged for ATP by the nucleotide exchange factor GrpE. It has been unclear how GrpE couples DnaK's nucleotide exchange with substrate release. Here we report a cryo-EM analysis of GrpE bound to an intact Mtb DnaK, revealing an asymmetric 1:2 DnaK-GrpE complex. The GrpE dimer ratchets to modulate both DnaK nucleotide-binding domain and the substrate-binding domain. We further show that the disordered GrpE N-terminus is critical for substrate release, and that the DnaK-GrpE interface is essential for protein folding activity both in vitro and in vivo. Therefore, the Mtb GrpE dimer allosterically regulates DnaK to concomitantly release ADP in the nucleotide-binding domain and substrate peptide in the substrate-binding domain.
Organizational Affiliation:
Department of Structural Biology, Van Andel Institute, Grand Rapids, MI, USA.