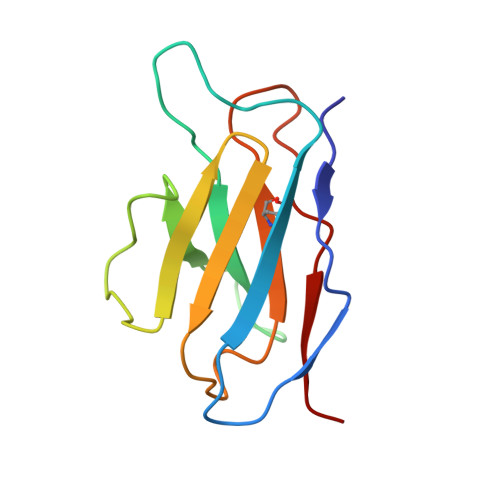
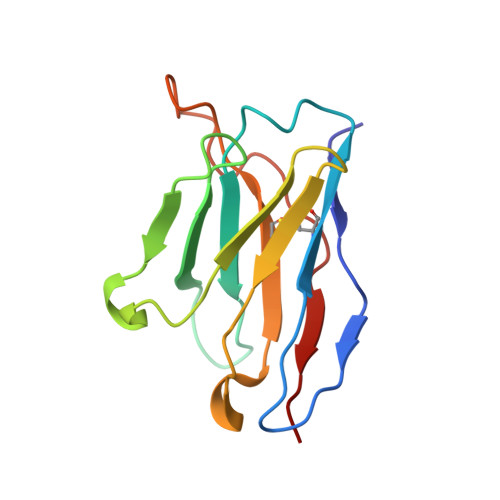
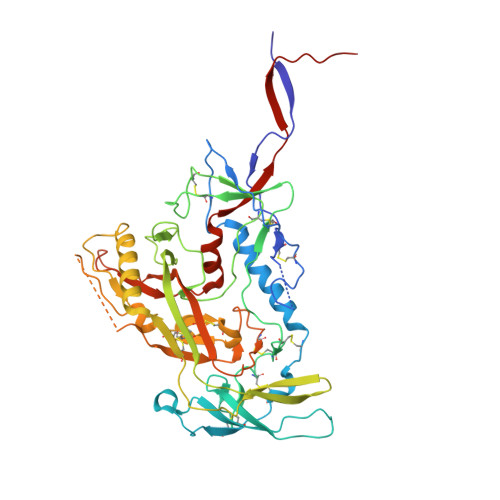
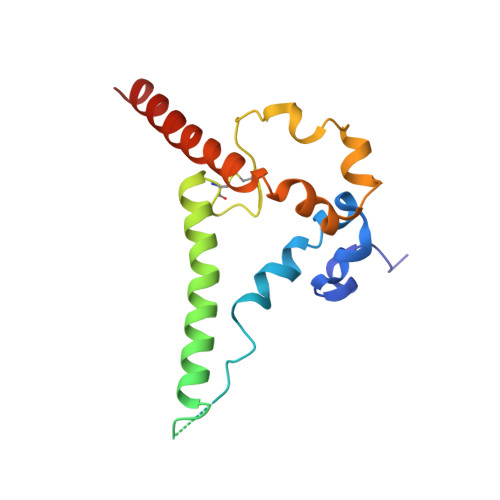
Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
Sastry, M., Changela, A., Gorman, J., Xu, K., Chuang, G.Y., Shen, C.H., Cheng, C., Geng, H., O'Dell, S., Ou, L., Rawi, R., Reveiz, M., Stewart-Jones, G.B.E., Wang, S., Zhang, B., Zhou, T., Biju, A., Chambers, M., Chen, X., Corrigan, A.R., Lin, B.C., Louder, M.K., McKee, K., Nazzari, A.F., Olia, A.S., Parchment, D.K., Sarfo, E.K., Stephens, T., Stuckey, J., Tsybovsky, Y., Verardi, R., Wang, Y., Zheng, C.Y., Chen, Y., Doria-Rose, N.A., McDermott, A.B., Mascola, J.R., Kwong, P.D.(2023) J Virol 97: e0160422-e0160422
- PubMed: 37098956
- DOI: https://doi.org/10.1128/jvi.01604-22
- Primary Citation of Related Structures:
8FR6, 8G85, 8G9W, 8G9X, 8G9Y, 8GAS - PubMed Abstract:
While neutralizing antibodies that target the HIV-1 fusion peptide have been elicited in mice by vaccination, antibodies reported thus far have been from only a single antibody class that could neutralize ~30% of HIV-1 strains. To explore the ability of the murine immune system to generate cross-clade neutralizing antibodies and to investigate how higher breadth and potency might be achieved, we tested 17 prime-boost regimens that utilized diverse fusion peptide-carrier conjugates and HIV-1 envelope trimers with different fusion peptides. We observed priming in mice with fusion peptide-carrier conjugates of variable peptide length to elicit higher neutralizing responses, a result we confirmed in guinea pigs. From vaccinated mice, we isolated 21 antibodies, belonging to 4 distinct classes of fusion peptide-directed antibodies capable of cross-clade neutralization. Top antibodies from each class collectively neutralized over 50% of a 208-strain panel. Structural analyses - both X-ray and cryo-EM - revealed each antibody class to recognize a distinct conformation of fusion peptide and to have a binding pocket capable of accommodating diverse fusion peptides. Murine vaccinations can thus elicit diverse neutralizing antibodies, and altering peptide length during prime can improve the elicitation of cross-clade responses targeting the fusion peptide site of HIV-1 vulnerability. IMPORTANCE The HIV-1 fusion peptide has been identified as a site for elicitation of broadly neutralizing antibodies, with prior studies demonstrating that priming with fusion peptide-based immunogens and boosting with soluble envelope (Env) trimers can elicit cross-clade HIV-1-neutralizing responses. To improve the neutralizing breadth and potency of fusion peptide-directed responses, we evaluated vaccine regimens that incorporated diverse fusion peptide-conjugates and Env trimers with variation in fusion peptide length and sequence. We found that variation in peptide length during prime elicits enhanced neutralizing responses in mice and guinea pigs. We identified vaccine-elicited murine monoclonal antibodies from distinct classes capable of cross-clade neutralization and of diverse fusion peptide recognition. Our findings lend insight into improved immunogens and regimens for HIV-1 vaccine development.
Organizational Affiliation:
Vaccine Research Center, National Institutes of Health, Bethesda, Maryland, USA.