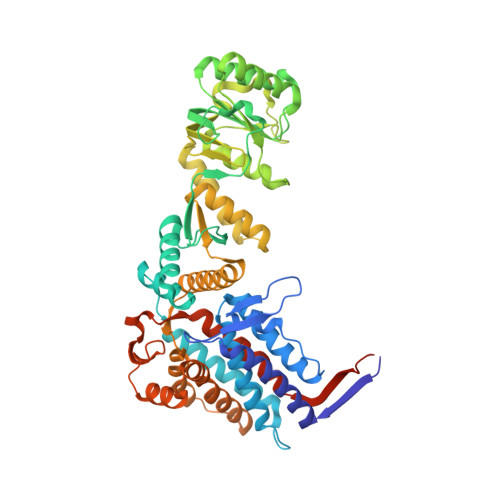
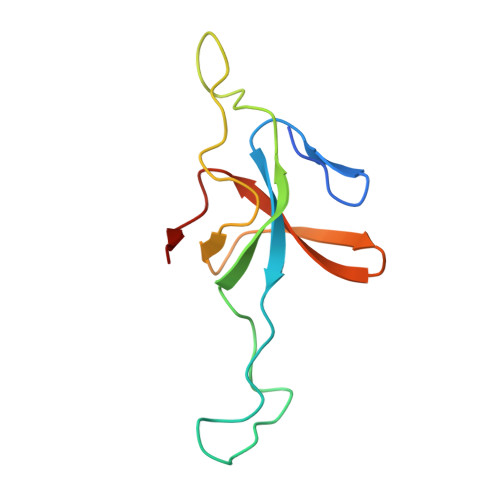
Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Braxton, J.R., Shao, H., Tse, E., Gestwicki, J.E., Southworth, D.R.(2023) bioRxiv
- PubMed: 37293102
- DOI: https://doi.org/10.1101/2023.05.15.540872
- Primary Citation of Related Structures:
8G7J, 8G7K, 8G7L, 8G7M, 8G7N, 8G7O - PubMed Abstract:
The mitochondrial chaperonin, mtHsp60, promotes the folding of newly imported and transiently misfolded proteins in the mitochondrial matrix, assisted by its co-chaperone mtHsp10. Despite its essential role in mitochondrial proteostasis, structural insights into how this chaperonin binds to clients and progresses through its ATP-dependent reaction cycle are not clear. Here, we determined cryo-electron microscopy (cryo-EM) structures of a hyperstable disease-associated mtHsp60 mutant, V72I, at three stages in this cycle. Unexpectedly, client density is identified in all states, revealing interactions with mtHsp60's apical domains and C-termini that coordinate client positioning in the folding chamber. We further identify a striking asymmetric arrangement of the apical domains in the ATP state, in which an alternating up/down configuration positions interaction surfaces for simultaneous recruitment of mtHsp10 and client retention. Client is then fully encapsulated in mtHsp60/mtHsp10, revealing prominent contacts at two discrete sites that potentially support maturation. These results identify a new role for the apical domains in coordinating client capture and progression through the cycle, and suggest a conserved mechanism of group I chaperonin function.
Organizational Affiliation:
Graduate Program in Chemistry and Chemical Biology; University of California, San Francisco; San Francisco, CA 94158, USA.