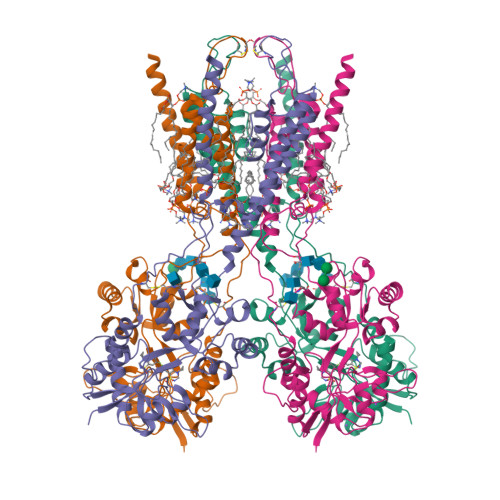
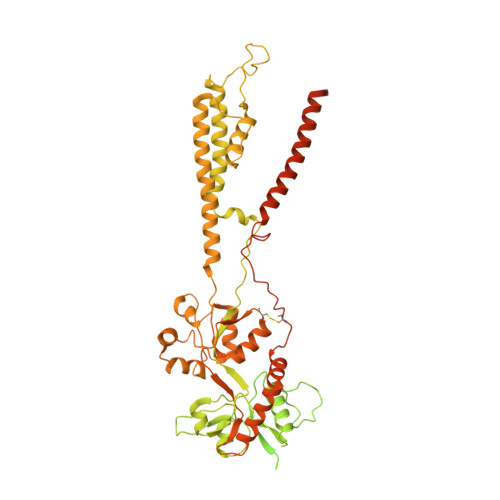
Positive and negative allosteric modulation of GluK2 kainate receptors by BPAM344 and antiepileptic perampanel.
Gangwar, S.P., Yen, L.Y., Yelshanskaya, M.V., Sobolevsky, A.I.(2023) Cell Rep 42: 112124-112124
- PubMed: 36857176
- DOI: https://doi.org/10.1016/j.celrep.2023.112124
- Primary Citation of Related Structures:
8FWQ, 8FWR, 8FWS, 8FWT, 8FWU, 8FWV, 8FWW - PubMed Abstract:
Kainate receptors (KARs) are a subtype of ionotropic glutamate receptors that control synaptic transmission in the central nervous system and are implicated in neurological, psychiatric, and neurodevelopmental disorders. Understanding the regulation of KAR function by small molecules is essential for exploring these receptors as drug targets. Here, we present cryoelectron microscopy (cryo-EM) structures of KAR GluK2 in complex with the positive allosteric modulator BPAM344, competitive antagonist DNQX, and negative allosteric modulator, antiepileptic drug perampanel. Our structures show that two BPAM344 molecules bind per ligand-binding domain dimer interface. In the absence of an agonist or in the presence of DNQX, BPAM344 stabilizes GluK2 in the closed state. The closed state is also stabilized by perampanel, which binds to the ion channel extracellular collar sites located in two out of four GluK2 subunits. The molecular mechanisms of positive and negative allosteric modulation of KAR provide a guide for developing new therapeutic strategies.
Organizational Affiliation:
Department of Biochemistry and Molecular Biophysics, Columbia University, 650 West 168(th) Street, New York, NY 10032, USA.