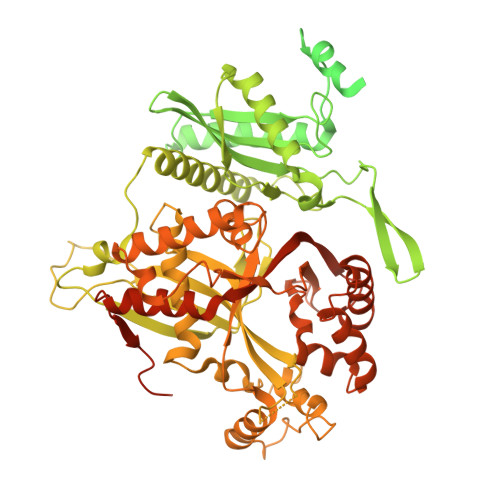
The phi PA3 phage nucleus is enclosed by a self-assembling 2D crystalline lattice.
Nieweglowska, E.S., Brilot, A.F., Mendez-Moran, M., Kokontis, C., Baek, M., Li, J., Cheng, Y., Baker, D., Bondy-Denomy, J., Agard, D.A.(2023) Nat Commun 14: 927-927
- PubMed: 36807264
- DOI: https://doi.org/10.1038/s41467-023-36526-9
- Primary Citation of Related Structures:
8FNE, 8FV5 - PubMed Abstract:
To protect themselves from host attack, numerous jumbo bacteriophages establish a phage nucleus-a micron-scale, proteinaceous structure encompassing the replicating phage DNA. Bacteriophage and host proteins associated with replication and transcription are concentrated inside the phage nucleus while other phage and host proteins are excluded, including CRISPR-Cas and restriction endonuclease host defense systems. Here, we show that nucleus fragments isolated from ϕPA3 infected Pseudomonas aeruginosa form a 2-dimensional lattice, having p2 or p4 symmetry. We further demonstrate that recombinantly purified primary Phage Nuclear Enclosure (PhuN) protein spontaneously assembles into similar 2D sheets with p2 and p4 symmetry. We resolve the dominant p2 symmetric state to 3.9 Å by cryo-EM. Our structure reveals a two-domain core, organized into quasi-symmetric tetramers. Flexible loops and termini mediate adaptable inter-tetramer contacts that drive subunit assembly into a lattice and enable the adoption of different symmetric states. While the interfaces between subunits are mostly well packed, two are open, forming channels that likely have functional implications for the transport of proteins, mRNA, and small molecules.
Organizational Affiliation:
Department of Biochemistry, University of California San Francisco, San Francisco, CA, 94143, USA.